-
PDF
- Split View
-
Views
-
Cite
Cite
Amanda Ewart-Toland, Qi Dai, Yu-Tang Gao, Hiroki Nagase, Malcolm G. Dunlop, Susan M. Farrington, Rebecca A. Barnetson, Hoda Anton-Culver, David Peel, Argyrios Ziogas, Dongxin Lin, Xiaoping Miao, Tong Sun, Elaine A. Ostrander, Janet L. Stanford, Mariela Langlois, June M. Chan, Jinwei Yuan, Curtis C. Harris, Elise D. Bowman, Gary L. Clayman, Scott M. Lippman, J. Jack Lee, Wei Zheng, Allan Balmain, Aurora- A/STK15 T + 91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types , Carcinogenesis, Volume 26, Issue 8, August 2005, Pages 1368–1373, https://doi.org/10.1093/carcin/bgi085
Close - Share Icon Share
Abstract
STK15 (Aurora-A) is a serine/threonine kinase involved in mitotic chromosomal segregation. A genetic variant in STK15T + 91A (resulting in the amino acid substitution F31I) is associated with increased aneuploidy in colon tumors and cell transformation in vitro . Since this polymorphism plays a role in mitotic control—a process critical for all cancer types—we conducted association analyses for risk of cancer development of the colon, breast, prostate, skin, lung and esophagus in 10 independent case–control populations. We carried out a meta-analysis of these 10 case–control studies together with 5 additional published studies for a total of 9549 cases of breast, colon, ovarian, prostate, lung, esophageal and non-melanoma skin cancer and 8326 population or hospital-based controls. Meta-analysis of three colorectal cancer studies showed an increased risk in T + 91A homozygotes (OR = 1.50; 95% CI of 1.14–1.99). Meta-analysis of four breast cancer studies showed increased risk for T + 91A homozygotes (OR = 1.35, 95% CI of 1.12–1.64). The results of the multiple cancer type meta-analysis for all 15 studies combined were significant for cancer risk in both homozygotes and heterozygotes. The T + 91A heterozygotes show an OR of 1.10 (95% CI of 1.03–1.18, P -value = 0.006) and the T + 91A homozygotes show an OR of 1.40 (95% CI of 1.22–1.59, P -value <0.001) for cancer risk. These results confirm that the STK15T + 91A variant is a low penetrance cancer susceptibility allele affecting multiple cancer types, and provide genetic evidence from large-scale human population studies that genetic stability at the chromosome level is an important determinant of cancer susceptibility. The data also underline the advantages of comparative association studies involving study populations from different ethnic groups for determination of disease risk.
Introduction
Previous studies using a combined mouse/human strategy for the identification of cancer susceptibility genes led to the identification of a variant in human STK15T + 91A (F31I) that is preferentially amplified and associated with the degree of aneuploidy in human colon tumors ( 1 ). The Ile31 variant transforms Rat1 cells more efficiently than the Phe31 variant. Yeast two-hybrid screens using the two forms of STK15 as bait identified UBE2N as a preferential binding partner of the Phe31 variant, but not the Ile31 variant. This interaction leads to colocalization of UBE2N and STK15 at the centrosomes during mitosis.
STK15 shows copy number gain or overexpression in a number of human carcinomas and cell lines including those of the breast, ovary, stomach, endometrium, pancreas, prostate, bladder and colon ( 2 – 11 ). Increased expression of STK15 is associated with chromosomal instability in breast cancer ( 12 ) and aneuploidy in gastric cancers ( 4 ). Because of the functional evidence for a role of the T + 91A variant and the importance of STK15 in control of mitosis and chromosomal segregation, processes central to development of multiple cancer types, we conducted a meta-analysis with pooled data from 15 studies to determine cancer risk associated with the Ile31 ( T + 91A ) variant in human populations.
Materials and methods
Populations used in study
Written informed consent was obtained from all study participants according to local guidelines. The relevant committees on human research and institutional review boards approved this research proposal. Fifteen independent populations were used in this meta-analysis. We generated new STK15T + 91A genotype data for 10 of those populations. Five additional populations were published for STK15T + 91A cancer risk at the time of analysis.
Colon cancer. We used three colon cancer case–control sets in this analysis. The colon cancer study from the University of Edinburgh, Scotland, consists of prospectively collected population-wide extreme early onset cancers ( n = 1425) as well as retrospectively ( n = 250) and prospectively identified population-wide cases arising under the age of 80 ( n = 1038). Controls were collected population-wide and were roughly matched based on age, sex and area of residence. Of the participants, >99% were of Caucasian ancestry. The University of California Irvine (UCI) colon cancer case–control group consisted of 344 cases ascertained through the population-based cancer registries, Cancer Surveillance Program of Orange County and the San Diego Imperial Organization for Cancer Control, and 448 controls ascertained through random digit dialing ( 13 ). Of the UCI cases, 86% were of Caucasian ancestry 5% were of Asian ancestry, 7% were of Hispanic ancestry and 1.5% were of African American ancestry. The breakdown of ethnicities in the UCI controls (also used in the UCI breast study) was 80% Caucasian, 4% Asian, 12% Hispanic and 1% African American. The third colorectal cancer case–control population consisted of 283 cases and matched controls from Bejing, China. Cases were patients with primary colorectal cancer recruited from January 1997 to June 2003 at the Cancer Hospital, Chinese Academy of Medical Sciences (CAMS, Beijing City). Cases were newly diagnosed, histologically confirmed and previously untreated (by radiotherapy or chemotherapy) incident cases. There were no age, sex or histological restrictions. Exclusion criteria included previous cancer, metastasized cancer from other organs, previous radiotherapy or chemotherapy. Senior pathologists at the hospital determined histological types of the cancer by postoperative histopathological examination or biopsy via colonoscopy. Controls ( n = 283) consisted of a subset of controls used for the CAMS esophageal cancer study and were matched to the cases based on age (within 5 years) and gender using SAS 6.12. All participants were of Han Chinese ancestry.
Breast cancer. Four case–control breast cancer populations were used in this analysis. The UCI breast cancer case–control study consisted of 898 breast cancer cases ascertained through the population-based cancer registry of the Cancer Surveillance Program of Orange County ( 14 ) and 448 population-based controls (same as used for the colon cancer study). Of the UCI breast cases, 89% were of Caucasian ancestry, 4% Asian ancestry and 6% African American ancestry. The Shanghai Breast Cancer Study (SBCS; 15 , 16 ) consisted of breast cancer cases ( n = 1102) with age frequency matched (within 5 years) community controls ( n = 1186). Controls included women with benign breast disease. Data from two additional breast cancer case–control studies were published for the STK15 genotypes prior to final analyses here. The US-based breast cancer study consisted of 940 breast cancer cases identified through a registry and 830 population-based controls ( 17 ). All women were of Caucasian ancestry. The published Han Chinese breast cancer study consisted of 520 hospital-based breast cancer cases and 520 population-based controls matched on age ( 18 ). All participants were of Han Chinese ancestry.
Prostate cancer. Two prostate cancer case–control sets were included in this meta-analysis. The first was a nested case–control set from the Physician's Health Study ( 19 ), a large cohort study ( n = 501 cases and 501 controls). Cases were primarily of Caucasian ancestry (96%) with the remaining 4% comprising Asian/Pacific Islander, African Americans, Hispanics and mixed ethnic background. Controls were also primarily of Caucasian ancestry (94%) with the remaining 6% comprising Asian/Pacific Islanders (1%), African Americans, Hispanics (3%), and mixed ethnic background. A second case–control set consisted of 559 cases of incident prostate cancer ascertained from the Seattle–Puget Sound Surveillance, Epidemiology and End Results Cancer Registry (SPS SEER Fred Hutchinson Cancer Research Center) with age matched population-based controls from King County, Washington ( n = 534) ( 20 , 21 ). SPS SEER cases were 95% Caucasian and 5% African American background, while SPS controls were 97% Caucasian and 3% African American.
Lung cancer. The one lung cancer case–control set consisted of population-based cases ( n = 414) of Caucasian (72.9%) or African American (27.1%) ancestry residing in Metropolitan Baltimore or the Maryland Eastern Shore ( 22 ). The controls included a set of hospital-based controls matched on age, gender, smoking and ethnicity (32% African American, 68% Caucasian, n = 203) and a set of population-based controls matched on age, gender and ethnicity (41.4% African American, 58.6% Caucasian, n = 264).
Non-melanoma skin. Hospital-based case patients ( n = 236) were recruited from The University of Texas M.D. Anderson Cancer Center (Houston, TX). All patients with either newly diagnosed or surgically treated, histopathologically confirmed non-melanoma skin cancer between July 1996 and June 2001 were eligible. Cancer-free control subjects ( n = 182) were recruited from among genetically unrelated clinic visitors and were individually matched to the case patients by age (within 5 years), sex and ethnicity. The exclusion criteria for case patients were prior chemotherapy or radiation therapy. The exclusion criteria for all study subjects were prior cancer (except for non-melanoma skin cancer for the case patients) and any blood transfusion in the 6 months prior to recruitment. Participation rate was >90% among both case and control subjects. The majority of participants in this study were of Caucasian ancestry.
Esophageal cancer. One esophageal cancer case–control set was tested in this analysis ( 23 ). Cases were prospectively recruited from the Cancer Hospital, Chinese Academy of Medical Sciences (CAMS). Population-based controls were matched on sex and age (±5 years). All cases and controls were of Chinese ancestry.
Ovarian cancer. Three ovarian cancer case–control populations from a published meta-analysis of the risk of STK15 polymorphisms and ovarian cancer were used ( 24 ). All three studies consisted of Caucasian women.
Genotyping analyses
Three primary genotyping methods were used: allelic discrimination assays using the ABI PRISM 7700 or 7900 sequence detection systems (Applied Biosystems, Foster City, CA), single base extension method (ABI SnAPSHOT from Applied Biosystems) and restriction fragment length polymorphism using the enzyme ApoI, which recognizes the A allele ( 23 ). Allelic discrimation assays using ABI PRISM 7700 or 7900 sequence detection systems were conducted according to recommendations by Applied Biosystems. Primer and probe sequences and conditions are available upon request. The ABI SnAPSHOT single base extensions were carried out according to manufacturer conditions. Primer sequences are available on request. Genotypes were assessed in a blinded method for the majority of studies and were independently reviewed by at least two researchers.
Statistical analyses
Deviation of the genotype frequencies from those expected under Hardy–Weinberg equilibrium was assessed in the controls by χ 2 tests. Genotype frequencies in cases and controls were compared by χ 2 tests. The genotypic specific cancer risks were estimated as odds ratios (OR) with associated 95% confidence limits by unconditional logistic regression. The meta-analysis was conducted using multiple logistic regression analysis through the STATA software package ( 25 ). Population attributable risk was calculated using the equation P (RR − 1)/[1 + P (RR − 1)] in which P is the proportion of population-based controls exposed to the genotype ( T + 91A heterozygotes or homozygotes) and the relative risk was estimated using odds ratios calculated in the meta-analysis.
Results
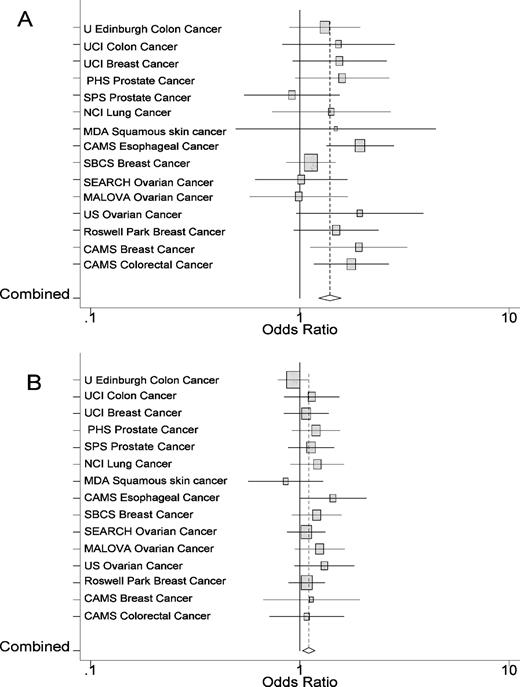
Based on genetic and functional evidence for STK15 / Stk6 in both human and mouse cancer, we genotyped the T + 91A variant to test for cancer risk in 10 cancer case–control populations. These populations had not previously been studied for this variant and included cancers of the prostate, colon, breast, lung, skin and esophagus ( Table I ) for a total of 6695 cases and 5012 control genotypes. Most of the case–control sets did not independently reach significance for cancer risk for homozygosity of the T + 91A polymorphism. However, 9 of the 10 studies showed trends or borderline significance for increased risk of the rare homozygote AA ( Table II , Figure 1A ). None of the sets differed significantly from expected allele frequencies by Hardy–Weinburg equilibrium tests in cases or controls.
Estimated OR with 95% CI for cancer risk odds ratios associated with homozygosity ( A ) and heterozygosity of STK15T + 91A ( B ) in all 15 studies. The area of each square is proportional to the variance of the log OR. The combined OR and 95% CI is denoted as a diamond. The combined OR is indicated as a dotted vertical line.
Studies tested for T+91A cancer association and included in meta-analysis
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Breast | UC Irvine (14) | Case–control | 898 | 448 a | US Californian |
| Shanghai Breast Cancer Study (SBCS;15) | Case–control | 1102 | 1186 | Chinese | |
| Colon | UC Irvine (13) | Case–control | 344 | 448 a | US Californian |
| U Edinburgh, Scotland | Case–control | 1675 | 1038 | Caucasian (Scottish) | |
| Chinese Academy of Medical Sciences (CAMS) | Case–control | 283 | 283 b | Han Chinese | |
| Esophageal | CAMS (23) | Case–control | 656 | 656 | Han Chinese |
| Lung | Metropolitan Baltimore, NCI (22) | Case–hospital and population-based controls | 414 | 467 | Caucasian/African American |
| Non-melanoma skin | MD Anderson | Case–control | 236 | 182 | Caucasian |
| Prostate | Physicians Health Study (19) | Cohort-nested case–control | 501 | 501 | Caucasian |
| FHCRC (20,21) | Case–control | 586 | 534 | Caucasian | |
| 10 studies | 6695 | 5012 |
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Breast | UC Irvine (14) | Case–control | 898 | 448 a | US Californian |
| Shanghai Breast Cancer Study (SBCS;15) | Case–control | 1102 | 1186 | Chinese | |
| Colon | UC Irvine (13) | Case–control | 344 | 448 a | US Californian |
| U Edinburgh, Scotland | Case–control | 1675 | 1038 | Caucasian (Scottish) | |
| Chinese Academy of Medical Sciences (CAMS) | Case–control | 283 | 283 b | Han Chinese | |
| Esophageal | CAMS (23) | Case–control | 656 | 656 | Han Chinese |
| Lung | Metropolitan Baltimore, NCI (22) | Case–hospital and population-based controls | 414 | 467 | Caucasian/African American |
| Non-melanoma skin | MD Anderson | Case–control | 236 | 182 | Caucasian |
| Prostate | Physicians Health Study (19) | Cohort-nested case–control | 501 | 501 | Caucasian |
| FHCRC (20,21) | Case–control | 586 | 534 | Caucasian | |
| 10 studies | 6695 | 5012 |
The same set of controls was used for both the breast and colon cancer studies from UC Irvine in the individual risk assessments.
The controls used for the CAMS colorectal cancer study were a subset of those used in the esophageal study.
Studies tested for T+91A cancer association and included in meta-analysis
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Breast | UC Irvine (14) | Case–control | 898 | 448 a | US Californian |
| Shanghai Breast Cancer Study (SBCS;15) | Case–control | 1102 | 1186 | Chinese | |
| Colon | UC Irvine (13) | Case–control | 344 | 448 a | US Californian |
| U Edinburgh, Scotland | Case–control | 1675 | 1038 | Caucasian (Scottish) | |
| Chinese Academy of Medical Sciences (CAMS) | Case–control | 283 | 283 b | Han Chinese | |
| Esophageal | CAMS (23) | Case–control | 656 | 656 | Han Chinese |
| Lung | Metropolitan Baltimore, NCI (22) | Case–hospital and population-based controls | 414 | 467 | Caucasian/African American |
| Non-melanoma skin | MD Anderson | Case–control | 236 | 182 | Caucasian |
| Prostate | Physicians Health Study (19) | Cohort-nested case–control | 501 | 501 | Caucasian |
| FHCRC (20,21) | Case–control | 586 | 534 | Caucasian | |
| 10 studies | 6695 | 5012 |
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Breast | UC Irvine (14) | Case–control | 898 | 448 a | US Californian |
| Shanghai Breast Cancer Study (SBCS;15) | Case–control | 1102 | 1186 | Chinese | |
| Colon | UC Irvine (13) | Case–control | 344 | 448 a | US Californian |
| U Edinburgh, Scotland | Case–control | 1675 | 1038 | Caucasian (Scottish) | |
| Chinese Academy of Medical Sciences (CAMS) | Case–control | 283 | 283 b | Han Chinese | |
| Esophageal | CAMS (23) | Case–control | 656 | 656 | Han Chinese |
| Lung | Metropolitan Baltimore, NCI (22) | Case–hospital and population-based controls | 414 | 467 | Caucasian/African American |
| Non-melanoma skin | MD Anderson | Case–control | 236 | 182 | Caucasian |
| Prostate | Physicians Health Study (19) | Cohort-nested case–control | 501 | 501 | Caucasian |
| FHCRC (20,21) | Case–control | 586 | 534 | Caucasian | |
| 10 studies | 6695 | 5012 |
The same set of controls was used for both the breast and colon cancer studies from UC Irvine in the individual risk assessments.
The controls used for the CAMS colorectal cancer study were a subset of those used in the esophageal study.
Genotypic breakdown of new studies
| Study . | Genotype . | Cases ( n ) . | Controls ( n ) . | OR . | 95% CI . |
|---|---|---|---|---|---|
| U Edinburgh colon | TT | 1031 | 630 | 1.00 | |
| AT | 558 | 368 | 0.93 | 0.79–1.09 | |
| AA | 86 | 40 | 1.31 | 0.89–1.94 | |
| UC Irvine colon | TT | 200 | 279 | 1.00 | |
| AT | 121 | 148 | 1.14 | 0.84–1.54 | |
| AA | 23 | 21 | 1.53 | 0.82–2.84 | |
| UC Irvine breast | TT | 533 | 279 | 1.00 | |
| AT | 303 | 148 | 1.07 | 0.84–1.37 | |
| AA | 62 | 21 | 1.55 | 0.92–2.59 | |
| PHS prostate | TT | 268 | 295 | 1.00 | |
| AT | 194 | 179 | 1.27 | 0.96–1.68 | |
| AA | 39 | 27 | 1.76 | 1.01–3.04 | |
| FHCRC prostate | TT | 348 | 329 | 1.00 | |
| AT | 208 | 174 | 1.13 | 0.88–1.45 | |
| AA | 30 | 31 | 0.91 | 0.54–1.55 | |
| NCI lung | TT | 266 | 322 | 1.00 | |
| AT | 127 | 127 | 1.21 | 0.90–1.63 | |
| AA | 21 | 18 | 1.41 | 0.74–2.71 | |
| MDA skin | TT | 151 | 112 | 1.00 | |
| AT | 75 | 65 | 0.86 | 0.57–1.29 | |
| AA | 10 | 5 | 1.48 | 0.49–4.46 | |
| CAMS esophageal ( 23 ) | TT | 58 | 91 | 1.00 | |
| AT | 290 | 316 | 1.44 | 1.00–2.08 | |
| AA | 308 | 249 | 1.94 | 1.34–2.81 | |
| SBCS breast (16) | TT | 121 | 149 | 1.00 | |
| AT | 491 | 503 | 1.20 | 0.92–1.57 | |
| AA | 490 | 534 | 1.13 | 0.86–1.48 | |
| CAMS colorectal | TT | 30 | 42 | 1.00 | |
| AT | 111 | 137 | 1.13 | 0.67–1.93 | |
| AA | 142 | 104 | 1.91 | 1.12–3.26 |
| Study . | Genotype . | Cases ( n ) . | Controls ( n ) . | OR . | 95% CI . |
|---|---|---|---|---|---|
| U Edinburgh colon | TT | 1031 | 630 | 1.00 | |
| AT | 558 | 368 | 0.93 | 0.79–1.09 | |
| AA | 86 | 40 | 1.31 | 0.89–1.94 | |
| UC Irvine colon | TT | 200 | 279 | 1.00 | |
| AT | 121 | 148 | 1.14 | 0.84–1.54 | |
| AA | 23 | 21 | 1.53 | 0.82–2.84 | |
| UC Irvine breast | TT | 533 | 279 | 1.00 | |
| AT | 303 | 148 | 1.07 | 0.84–1.37 | |
| AA | 62 | 21 | 1.55 | 0.92–2.59 | |
| PHS prostate | TT | 268 | 295 | 1.00 | |
| AT | 194 | 179 | 1.27 | 0.96–1.68 | |
| AA | 39 | 27 | 1.76 | 1.01–3.04 | |
| FHCRC prostate | TT | 348 | 329 | 1.00 | |
| AT | 208 | 174 | 1.13 | 0.88–1.45 | |
| AA | 30 | 31 | 0.91 | 0.54–1.55 | |
| NCI lung | TT | 266 | 322 | 1.00 | |
| AT | 127 | 127 | 1.21 | 0.90–1.63 | |
| AA | 21 | 18 | 1.41 | 0.74–2.71 | |
| MDA skin | TT | 151 | 112 | 1.00 | |
| AT | 75 | 65 | 0.86 | 0.57–1.29 | |
| AA | 10 | 5 | 1.48 | 0.49–4.46 | |
| CAMS esophageal ( 23 ) | TT | 58 | 91 | 1.00 | |
| AT | 290 | 316 | 1.44 | 1.00–2.08 | |
| AA | 308 | 249 | 1.94 | 1.34–2.81 | |
| SBCS breast (16) | TT | 121 | 149 | 1.00 | |
| AT | 491 | 503 | 1.20 | 0.92–1.57 | |
| AA | 490 | 534 | 1.13 | 0.86–1.48 | |
| CAMS colorectal | TT | 30 | 42 | 1.00 | |
| AT | 111 | 137 | 1.13 | 0.67–1.93 | |
| AA | 142 | 104 | 1.91 | 1.12–3.26 |
Genotypic breakdown of new studies
| Study . | Genotype . | Cases ( n ) . | Controls ( n ) . | OR . | 95% CI . |
|---|---|---|---|---|---|
| U Edinburgh colon | TT | 1031 | 630 | 1.00 | |
| AT | 558 | 368 | 0.93 | 0.79–1.09 | |
| AA | 86 | 40 | 1.31 | 0.89–1.94 | |
| UC Irvine colon | TT | 200 | 279 | 1.00 | |
| AT | 121 | 148 | 1.14 | 0.84–1.54 | |
| AA | 23 | 21 | 1.53 | 0.82–2.84 | |
| UC Irvine breast | TT | 533 | 279 | 1.00 | |
| AT | 303 | 148 | 1.07 | 0.84–1.37 | |
| AA | 62 | 21 | 1.55 | 0.92–2.59 | |
| PHS prostate | TT | 268 | 295 | 1.00 | |
| AT | 194 | 179 | 1.27 | 0.96–1.68 | |
| AA | 39 | 27 | 1.76 | 1.01–3.04 | |
| FHCRC prostate | TT | 348 | 329 | 1.00 | |
| AT | 208 | 174 | 1.13 | 0.88–1.45 | |
| AA | 30 | 31 | 0.91 | 0.54–1.55 | |
| NCI lung | TT | 266 | 322 | 1.00 | |
| AT | 127 | 127 | 1.21 | 0.90–1.63 | |
| AA | 21 | 18 | 1.41 | 0.74–2.71 | |
| MDA skin | TT | 151 | 112 | 1.00 | |
| AT | 75 | 65 | 0.86 | 0.57–1.29 | |
| AA | 10 | 5 | 1.48 | 0.49–4.46 | |
| CAMS esophageal ( 23 ) | TT | 58 | 91 | 1.00 | |
| AT | 290 | 316 | 1.44 | 1.00–2.08 | |
| AA | 308 | 249 | 1.94 | 1.34–2.81 | |
| SBCS breast (16) | TT | 121 | 149 | 1.00 | |
| AT | 491 | 503 | 1.20 | 0.92–1.57 | |
| AA | 490 | 534 | 1.13 | 0.86–1.48 | |
| CAMS colorectal | TT | 30 | 42 | 1.00 | |
| AT | 111 | 137 | 1.13 | 0.67–1.93 | |
| AA | 142 | 104 | 1.91 | 1.12–3.26 |
| Study . | Genotype . | Cases ( n ) . | Controls ( n ) . | OR . | 95% CI . |
|---|---|---|---|---|---|
| U Edinburgh colon | TT | 1031 | 630 | 1.00 | |
| AT | 558 | 368 | 0.93 | 0.79–1.09 | |
| AA | 86 | 40 | 1.31 | 0.89–1.94 | |
| UC Irvine colon | TT | 200 | 279 | 1.00 | |
| AT | 121 | 148 | 1.14 | 0.84–1.54 | |
| AA | 23 | 21 | 1.53 | 0.82–2.84 | |
| UC Irvine breast | TT | 533 | 279 | 1.00 | |
| AT | 303 | 148 | 1.07 | 0.84–1.37 | |
| AA | 62 | 21 | 1.55 | 0.92–2.59 | |
| PHS prostate | TT | 268 | 295 | 1.00 | |
| AT | 194 | 179 | 1.27 | 0.96–1.68 | |
| AA | 39 | 27 | 1.76 | 1.01–3.04 | |
| FHCRC prostate | TT | 348 | 329 | 1.00 | |
| AT | 208 | 174 | 1.13 | 0.88–1.45 | |
| AA | 30 | 31 | 0.91 | 0.54–1.55 | |
| NCI lung | TT | 266 | 322 | 1.00 | |
| AT | 127 | 127 | 1.21 | 0.90–1.63 | |
| AA | 21 | 18 | 1.41 | 0.74–2.71 | |
| MDA skin | TT | 151 | 112 | 1.00 | |
| AT | 75 | 65 | 0.86 | 0.57–1.29 | |
| AA | 10 | 5 | 1.48 | 0.49–4.46 | |
| CAMS esophageal ( 23 ) | TT | 58 | 91 | 1.00 | |
| AT | 290 | 316 | 1.44 | 1.00–2.08 | |
| AA | 308 | 249 | 1.94 | 1.34–2.81 | |
| SBCS breast (16) | TT | 121 | 149 | 1.00 | |
| AT | 491 | 503 | 1.20 | 0.92–1.57 | |
| AA | 490 | 534 | 1.13 | 0.86–1.48 | |
| CAMS colorectal | TT | 30 | 42 | 1.00 | |
| AT | 111 | 137 | 1.13 | 0.67–1.93 | |
| AA | 142 | 104 | 1.91 | 1.12–3.26 |
If being homozygous for the T + 91A variant does not increase risk of developing cancer, one would expect that most of these studies would show no trends towards increase in risk, and one might expect that some studies would show a decrease in risk. Because only one of the studies we conducted (Prostate Cancer, Fred Hutchinson Cancer Research Center) showed an OR ∼1.0, and all of the other studies showed a trend of an OR of 1.1–1.9, we hypothesized that the T + 91A variant did contribute to cancer risk, although at a very modest level. Based on these initial studies, we estimated that the STK15T + 91A homozyogotes have an OR of ∼1.3. It can be calculated that to conform to the recommended criteria ( 26 ) for significance of an association study for an allele with a frequency of 21% (in Caucasian populations) and a typical OR of 1.3 for a low penetrance susceptibility gene, a sample size of ∼11 000 cases and 11 000 matched controls would be required to give 90% power at P = 0.0001 using a specific test of the recessive hypothesis. In Chinese populations the Ile31 homozygotes have a frequency of ∼44%, so a set of ∼1500 cases and controls were needed to reach a similar degree of power ( 14 , 21 ). We did not have access to a case–control set as large as this for any one type of cancer from either population. Since we did not see evidence of heterogeneity of risk for the heterozygote or homozygotes between the types of cancers tested ( P -value = 0.69; P -value = 0.37, respectively) and there is evidence for a role of STK15 in a variety of cancers, we decided to test the STK15T + 91A variant for general cancer risk by combining all available data into a large meta-analysis.
At the time of meta-analysis, five additional association studies for cancer risk and the T + 91A polymorphism in STK15 were published or ‘in press’ ( Table III ). We, therefore, pooled our genotyping data with additional data from all STK15 studies that we were aware of at the time of analysis ( Tables II and III ). A few additional groups declined to participate in this study because their independent results had not been published. Two additional studies published in abstract form only, that looked at both familial and population-based prostate cancer ( 27 ) and breast cancer ( 28 ), showed no large differences in allele frequencies between the cases and controls. The abstracts did not contain the breakdown of individuals in the three genotypic groups and are, therefore, not included in this study. Since we were using all of the studies for which we had access to data and because most were not published, it is likely that publication biases did not affect our analyses or conclusions.
Previously published studies included in meta-analysis
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Ovary ( 24 ) | SEARCH (UK) | Case–control | 752 | 843 | Caucasian/Anglian |
| Ovary ( 24 ) | MALOVA (Denmark) | Case–control | 334 | 723 | Caucasian/Danish |
| Ovary ( 24 ) | USA | Case–control | 308 | 398 | Caucasian/US |
| Breast ( 17 ) | Vanderbilt–Roswell Park | Case–control | 940 | 830 | Caucasian/US |
| Breast ( 18 ) | CAMS, China | Case–control | 520 | 520 | Han Chinese |
| 5 studies | 2854 | 3314 |
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Ovary ( 24 ) | SEARCH (UK) | Case–control | 752 | 843 | Caucasian/Anglian |
| Ovary ( 24 ) | MALOVA (Denmark) | Case–control | 334 | 723 | Caucasian/Danish |
| Ovary ( 24 ) | USA | Case–control | 308 | 398 | Caucasian/US |
| Breast ( 17 ) | Vanderbilt–Roswell Park | Case–control | 940 | 830 | Caucasian/US |
| Breast ( 18 ) | CAMS, China | Case–control | 520 | 520 | Han Chinese |
| 5 studies | 2854 | 3314 |
Previously published studies included in meta-analysis
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Ovary ( 24 ) | SEARCH (UK) | Case–control | 752 | 843 | Caucasian/Anglian |
| Ovary ( 24 ) | MALOVA (Denmark) | Case–control | 334 | 723 | Caucasian/Danish |
| Ovary ( 24 ) | USA | Case–control | 308 | 398 | Caucasian/US |
| Breast ( 17 ) | Vanderbilt–Roswell Park | Case–control | 940 | 830 | Caucasian/US |
| Breast ( 18 ) | CAMS, China | Case–control | 520 | 520 | Han Chinese |
| 5 studies | 2854 | 3314 |
| Cancer type . | Study . | Type . | Cases ( n ) . | Controls ( n ) . | Ethnicity . |
|---|---|---|---|---|---|
| Ovary ( 24 ) | SEARCH (UK) | Case–control | 752 | 843 | Caucasian/Anglian |
| Ovary ( 24 ) | MALOVA (Denmark) | Case–control | 334 | 723 | Caucasian/Danish |
| Ovary ( 24 ) | USA | Case–control | 308 | 398 | Caucasian/US |
| Breast ( 17 ) | Vanderbilt–Roswell Park | Case–control | 940 | 830 | Caucasian/US |
| Breast ( 18 ) | CAMS, China | Case–control | 520 | 520 | Han Chinese |
| 5 studies | 2854 | 3314 |
To determine if there was a statistically significant increase in risk for any one type of cancer, we conducted ‘mini’ meta-analyses for the two cancer types, breast and colon, for which we had three or more independent studies. Neither the breast nor the colorectal cancer studies showed evidence for an increase in risk of the heterozygotes. (Colorectal cancer: OR = 0.98, 95% CI of 0.85–1.13; P -value = 0.8; breast cancer; OR = 1.10, 95% CI of 0.97–1.25; P -value = 0.13.) However in our colon cancer meta-analysis, we found an OR of 1.50 for the T + 91A homozygotes (95% CI of 1.14–1.99, P -value = 0.004). Our breast cancer meta-analysis also showed an increase in cancer risk for the T + 91A homozygotes (OR = 1.35, 95% CI of 1.12–1.64; P -value = 0.002) when the genotypes from the four breast cancer studies were combined.
When the results of all available studies (15 case–control sets; 9549 cases and 8326 controls) were combined in a large meta-analysis, we found an OR of 1.4 for the T + 91A homozygotes (95% CI of 1.22–1.59, P -value < 0.001, Figure 1A ). In addition, we saw a very modest but significant increase in risk in the T + 91A heterozygotes (OR = 1.10, 95% CI 1.03–1.18, P -value = 0.006, Figure 1B ). These results confirm that the STK15T + 91A polymorphism is a low penetrance cancer susceptibility allele important in multiple cancer types.
Discussion
These studies confirm the STK15 T + 91A variant as a low penetrance cancer susceptibility allele. What is striking is that several independent case–control studies have shown similar trends for increased risk of the T + 91A homozygotes (OR of 1.1–1.5), but the majority by themselves did not reach statistical significance. A meta-analysis using several thousand cases and controls was needed to reach an acceptable level of significance. In addition, the very modest increase in risk for heterozygote carriers was only seen in the large meta-analysis and not in individual studies or smaller meta-analyses. Most of these independent studies were not small with an average number of cases and controls in each independent collection of ∼650. It is likely that many cancer susceptibility genes will show similar modest increases in risk that will only reach statistical significance in very large studies or meta-analyses.
This work represents one of the few meta-analyses to date looking at a variant for risk in multiple cancer types. Several variants have been implicated in cancer risk in multiple cancer types, such as HRAS ( 29 ) and CHEK2 ( 30 ), but few multi-cancer studies have been conducted. The only other recent study we were able to find that assessed a variant for cancer risk in multiple cancer types examined the TGFBRI* 6A variant in 2438 mixed cancer cases and 1846 controls ( 31 ). Homozygote carriers in this study had an increased risk OR of 2.53. It is likely that some variants reported in the literature to be associated with a specific type of cancer risk will be general cancer susceptibility factors and that meta-analyses of multiple cancer types will lead to a better understanding of overall risk.
As gene discovery in disease moves from identification of high-penetrance mutations with Mendelian inheritance to identification of low-penetrance variants that in part contribute to complex traits, the ease of proving causality decreases rapidly. Association studies, the main workhorse for identification of low penetrance disease alleles, are fraught with problems. The results of association studies for diseases ranging from diabetes to cancer risk are often not replicated in completely independent populations. The majority of the independent studies shown here were not significant on their own, in part due to the low risk associated with homozygosity of the T + 91A allele and the low frequency of homozygotes in Caucasian populations. A magnitude of 1.40 for the OR is not unexpected for the type of risk we might expect to see with a low penetrance allele, but makes replication difficult unless large numbers of cases and controls are used. This variant is not the first to be implicated in disease with such a low increase in risk. The PPARγ Pro12Ala polymorphism, e.g. is associated with a 1.25-fold increase in risk for diabetes with the most common allele ( 32 ). Original studies of this variant were positive, but subsequent studies failed to confirm the initial findings. It was not until a meta-analysis of over 3000 individuals was performed that the P -value for risk was considered significant.
Our study also demonstrates the value of comparative association studies in different ethnic groups. We saw large allele frequency differences between Caucasians and Chinese populations. In Caucasian populations the overall T + 91A allele frequency for cases and controls was 22.9 and 21.5%, respectively. In contrast, in the Chinese populations studied, the T + 91A allele frequency was 68.3% in cases and 64.2% in controls. Homozygotes for the T + 91A allele are ∼9 times more common in Chinese than in Caucasians. Even though the risk for cancer is similar in the two populations, the population attributable risk (PAR) is very different due to the large differences in allele frequency of the T + 91A variant. The PAR is estimated to be only 1.9% for T + 91A homozygotes and 3.2% for heterozygotes in Caucasian populations, but 14.2% for T + 91A homozygotes and 4.4% for heterozygotes in Chinese populations.
To contrast the differences we observed between populations, the overall difference in frequency of the Ile31 ( T + 91A ) between cases and controls within a population did not vary much (1.4% in Caucasians, 4.1% in Chinese). Therefore, DNA pooling methodologies that generally have standard errors of 2–5% for detection of allele frequency differences would not likely have identified this variant as important for cancer risk, particularly in the Caucasian populations ( 33 ).
We saw a similar magnitude of risk conferred by the allele in both the Caucasian and Chinese populations, but since the allele frequency is much higher in Chinese populations, fewer cases and controls were needed to reach similar degrees of significance. One future strategy that might be employed for these types of studies is to conduct follow-up studies in populations with a higher frequency of the variant allele when a marginal effect of a variant in one population is detected.
The T + 91A variant only contributes a modest increase in risk for cancer. Therefore, it does not have clinical relevance on its own. However, this variant may act synergistically or additively with other genetic variants to increase cancer risk. The identification of these predicted genetic interacting factors is crucial for further stratifying risk within populations. The genetic data presented here, together with the functional evidence for the role of STK15 in cell transformation, support this gene as a suitable genetic target for therapeutic or preventative drug development.
Identification of candidate genes for cancer risk is a major focus of research efforts. Several reports have shown that variants in DNA repair genes are implicated in increased risk in a variety of cancer types ( 34 , 35 ). The roles of instability ( 36 , 37 ) and aneuploidy ( 38 , 39 ) in cancer development have been widely discussed. Here, we show that a variation in a gene with a role in chromosomal stability is important in cancer risk. Our data implicate genetic control of chromosomal instability and aneuploidy as important factors in human cancer susceptibility.
We thank Drs Bruce Ponder, Paul Pharoah and Doug Easton for thoughtful discussions and sharing of data. The following funding bodies supported this work: the UCSF School of Medicine Research Evaluation and Allocation Committee fund, the Stewart Trust, the UCSF Prostate Spore Grant CA 89520-01, the UCSF Prostate Cancer Center Award, the National Cancer Institute (NCI) P01-5P01CA68233, the Cancer Center Support Grant 5P30 CA16672, the NCI Mouse Models of Human Cancer Consortium Supplement CA84244-03 S1, the National Institutes of Health (NIH) 1 P50 CA89520, the Medical Research Council G0000657-53203, the Cancer Research UK C348/A3758, the Chief Scientist's Office K/OPR/2/2/D33, the NCI Grant CA56678, the NCI grant CA82664 and the NCI contract N01-CN-05230. A.B. acknowledges the support of the Barbara Bass Bakar endowed chair in this work.
Conflict of Interest Statement : None declared.
References
Ewart-Toland,A., Briassouli,P., de Koning,J.P., Mao,J.H., Yuan,J., Chan,F., MacCarthy-Morrogh,L., Ponder,B.A., Nagase,H., Burn,J., Ball,S., Almeida,M., Linardopoulos,S. and Balmain,A. (
Sen,S., Zhou,H. and White,R.A. (
Bischoff,J.R., Anderson,L., Zhu,Y., Mossie,K., Ng,L., Souza,B., Schryver,B., Flanagan,P., Clairvoyant,F., Ginther,C., Chan,C.S., Novotny,M., Slamon,D.J. and Plowman,G.D. (
Sen,S., Zhou,H., Zhang,R.D. et al . (
Tanaka,T., Kimura,M., Matsunaga,K., Fukada,D., Mori,H. and Okano,Y. (
Tanner,M.M., Grenman,S., Koul,A., Johannsson,O., Meltzer,P., Pejovic,T., Borg,A. and Isola,J.J. (
Sakakura,C., Hagiwara,A., Yasuoka,R. et al . (
Bar-Shira,A., Pinthus,J.H., Rozovsk,U., Goldstein,M., Sellers,W.R., Yaron,Y., Eshhar,Z. and Orr-Urtreger,A. (
Watanabe,T., Imoto,I., Katahira,T., Hirasawa,A., Ishiwata,I., Emi,M., Takayama,M., Sato,A. and Inazawa,J. (
Li,D., Zhu,J., Firozi,P.F., Abbruzzese,J.L., Evans,D.B., Cleary,K., Friess,H. and Sen,S. (
Moreno-Bueno,G., Sanchez-Estevez,C., Cassia,R. et al . (
Miyoshi,Y., Iwao,K., Egawa,C. and Noguchi,S. (
Peel,D.J., Ziogas,A., Fox,A.F., Gildea,M., Laham,B., Clements,E., Kolodner,R.D. and Anton-Culver,H. (
Anton-Culver,H., Cohen,P.F., Gildea,M.E. and Ziogas,A. (
Gao,Y.T., Shu,X.O., Dai,Q. et al . (
Dai,Q., Shu,X-O., Cai,Q-Y,, Ewart-Toland,A., Wen,W-Q., Balmain,A., Gao,Y-T. and Zheng,W. (
Egan,K.M., Nagase,H., Newcomb,P.A., Trentham-Dietz,A., Titus-Ernstoff,L., Hampton,J.M., Kimura,M.T. and Nagase,H. (
Sun,T., Miao,X., Wang,J., Tan,W., Zhou,Y., Yu,C. and Lin,D. (
Chan,J.M., Stampfer,M.J., Ma,J., Gann,P., Gaziano,J.M., Pollak,M. and Giovannucci,E. (
Stanford,J.L., Noonan,E.A., Iwasaki,L., Kolb,S., Chadwick,R.B., Feng,Z. and Ostrander,E.A. (
Stanford,J.L., Wicklund,K.G., McKnight,B., Daling,J.R. and Brawer,M.K. (
Zheng,Y.L., Loffredo,C.A., Yu,Z.P. et al . (
Miao,X., Sun,T., Wang,Y., Zhang,X., Tan,W. and Lin,D. (
DiCioccio,R.A., Song,H., Waterfall,C. et al . (
Egger,M., Smith,G.D. and Altman,D.G. (
Dahlman,I., Eaves,I.A., Kosoy,R. et al . (
Wang,L., Elkins,D.A., McDonnell,S.K. et al . (
Olson,J.E., Liang,W., Hebbring,S., Vierknat,R.A., Fredericksen,Z.S., Pankratz,S., Couch,F.J., Thibodeau,S.N. and Sellers,T.A. (
Krontiris,T.G., Devlin,B., Karp,D.D., Robert,N.J. and Risch,N. (
Cybulski,C., Gorski,B., Huzarski,T. et al . (
Kaklamani,V.G., Hou,N., Bian,Y., Reich,J., Offit,K., Michel,L.S., Rubinstein,W.S., Rademaker,A. and Pasche,B. (
Altshuler,D., Hirschhorn,J.N., Klannemark,M. et al . (
Sham,P., Bader,J.S., Craig,I., O'Donovan,M. and Owen,M. (
Goode,E.L., Ulrich,C.M. and Potter,J.D. (
Loktionov,A. (
Sieber,O.M., Heinimann,K. and Tomlinson,I.P.M. (
Wang,Z., Cummins,J.M., Shen,D. et al . (
Sudbo,J., Kildal,W., Risberg,B., Koppang,H.S., Danielsen,H.E. and Reith,A. (
Author notes
1UCSF Comprehensive Cancer Center, University of California, San Francisco, CA, USA, 2Department of Medicine and Vanderbilt-Ingrim Cancer Center, School of Medicine, Vanderbilt University, Nashville TN 37232-8300, USA, 3Department of Epidemiology, Shanghai, Cancer Institute, Shanghai 200032, China, 4Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, NY, USA, 5Division of Oncology, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU, UK, 6Epidemiology Division, Department of Medicine, University of California, Irvine, Irvine, CA, USA, 7Department of Etiology and Carcinogenesis, Cancer Institute and Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, China, 8Divisions of Human Biology and Clinical Research and 9Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA, 10Department of Epidemiology, Department of Biostatistics and Department of Urology, University of California, San Francisco, CA, USA, 11Laboratory of Human Carcinogenesis, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA, 12Department of Head and Neck Surgery and Cancer Biology, 13Thoracic/Head and Neck Medical Oncology and Clinical Cancer Prevention, 14Biostatistics, M. D. Anderson Cancer Center, Houston, TX, USA and 15Department of Biochemistry and Biophysics (A.B), University of California San Francisco, San Francisco, CA 94115, USA