-
PDF
- Split View
-
Views
-
Cite
Cite
Kaisar A. Talukder, Bijay K. Khajanchi, M. Aminul Islam, Dilip K. Dutta, Zhahirul Islam, Ashrafus Safa, G. Y. Khan, Khorshed Alam, M. A. Hossain, Sarala Malla, S. K. Niyogi, Mustafizur Rahman, Haruo Watanabe, G. Balakrish Nair, David A. Sack, Genetic relatedness of ciprofloxacin-resistant Shigella dysenteriae type 1 strains isolated in south Asia, Journal of Antimicrobial Chemotherapy, Volume 54, Issue 4, October 2004, Pages 730–734, https://doi.org/10.1093/jac/dkh425
Close - Share Icon Share
Abstract
Objectives: The aim of the present study was to determine the clonal relationships of ciprofloxacin-resistant Shigella dysenteriae type 1 strains isolated from south Asia, and S. dysenteriae 1 strains associated with epidemics in 1978, 1984 and 1994.
Methods: The antimicrobial susceptibilities were examined by NCCLS methods. Molecular epidemiological characterization was performed by plasmid profiling, pulsed-field gel electrophoresis (PFGE) and mutation analysis of the quinolone resistance-determining region (QRDR) of gyrA by sequencing.
Results: Plasmid patterns of the current ciprofloxacin-resistant strains from India, Nepal and Bangladesh were very similar to those of the 1978, 1984 and 1994 epidemic isolates of S. dysenteriae 1, except for the presence of a new plasmid of ∼2.6 MDa, which was found in one recent ciprofloxacin-resistant strain isolated in Bangladesh. PFGE analysis showed that the ciprofloxacin-resistant strains isolated in Bangladesh, India and Nepal belonged to a PFGE type (type A), which was possibly related to that of the 1984 and 1994 clone of S. dysenteriae 1, but different from 1978 epidemic strains. The current ciprofloxacin-resistant strains belong to five subtypes (A3–A7), all of which were found in India, but in Bangladesh and Nepal, only A3 existed. Mutation analysis of the QRDR of gyrA revealed that amino acid substitutions at positions 83 and 87 of ciprofloxacin-resistant strains isolated in Bangladesh were similar to those of the strains isolated in Nepal, but different (at position 87) from ciprofloxacin-resistant strains isolated in India.
Conclusions: PFGE and mutation analysis of gyrA showed differences between the current ciprofloxacin-resistant S. dysenteriae 1 strains isolated in south Asia and those associated with epidemics in 1978, 1984 and 1994.
Introduction
Epidemic shigellosis caused by multidrug-resistant Shigella dysenteriae type 1 is a recurrent challenge in many parts of the developing world. Periodic lapses and emergence of strains resistant to new drugs have further complicated the treatment of infection caused by this pathogen. In Bangladesh, at least three large epidemics have been encountered since 19731 and each epidemic was associated with a high rate of mortality, particularly in children <4 years of age.2 In each epidemic, S. dysenteriae 1 strains emerged acquiring resistance to the latest drug of choice, together with other effective drugs, and this was evident in epidemics of 1973–1974, 1983–1984 and 1993–1994, where strains were resistant to ampicillin, sulfamethoxazole/trimethoprim and nalidixic acid, respectively.3–5 In the recent past, a decrease in prevalence of S. dysenteriae 1 has been observed in Bangladesh.1 During April–May 2002, only two strains of S. dysenteriae 1 were isolated in Dhaka, Bangladesh. These were resistant to ampicillin, sulfamethoxazole/trimethoprim and nalidixic acid, but were susceptible to ciprofloxacin,1 an effective drug for the treatment of Shigella infection. An upsurge of S. dysenteriae 1 resistant to ciprofloxacin and norfloxacin has recently been reported from eastern parts of India, and this resurgence was witnessed after a lapse of 14 years.6 From previous epidemics of S. dysenteriae 1, it has been demonstrated that the occurrence of an outbreak or emergence of resistance to new antibiotics in S. dysenteriae 1 in any country within the southeast Asia region forecasts the same episode in neighbouring countries. For instance, when an outbreak of nalidixic acid-resistant S. dysenteriae 1 occurred in 1988 in an eastern state of India,7 nalidixic-acid resistant S. dysenteriae 1 were sporadically isolated in Bangladesh until it emerged as an epidemic in 1993–1994.5
In this study, in order to ascertain its genesis, we compared the antimicrobial resistance pattern, genetic fingerprint and QRDR of gyrA of the current ciprofloxacin-resistant S. dysenteriae 1 strains isolated in south Asia, and the strains of S. dysenteriae 1 associated with the epidemics of 1978, 1984 and 1994.
Materials and methods
Bacterial strains
The origin and period of isolation of the strains in the present study are displayed in Table 1. Five strains of S. dysenteriae type 1 used in this study were isolated from patients attending the Dhaka and Matlab treatment centre operated by ICDDR, B; and three strains from Nepal were sent to us from the National Public Health Laboratory, Kathmandu, Nepal. Previous epidemic strains of S. dysenteriae 1 in Bangladesh and from India (Bombay and Kolkata) were collected from our stock; ciprofloxacin-resistant outbreak strains from Eastern India were gifted by the National Institute of Cholera and Enteric Diseases, Kolkata, India. Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 25923) were used as control strains for susceptibility studies.
Susceptibility testing
According to the guidelines of the National Committee for Clinical Laboratory Standards,8 antibiotic susceptibility testing was conducted by disc diffusion using commercially available antibiotic discs (Oxoid, Basingstoke, UK). The antibiotic discs used in this study were tetracycline (30 mg), ampicillin (10 mg), sulfamethoxazole/trimethoprim (25 mg), nalidixic acid (30 mg), ciprofloxacin (5 mg), norfloxacin (10 mg), ofloxacin (5 mg), mecillinam (25 mg), azithromycin (15 mg) and ceftriaxone (30 mg). The MIC of ciprofloxacin and nalidixic acid were determined by the Etest (AB Biodisk, Solna, Sweden).
Plasmid profile analysis
Plasmid DNA was prepared by the alkaline lysis method of Kado & Liu, with some modifications.9 The molecular weight of the unknown plasmid DNA was assessed by comparison with the mobilities of plasmids of known molecular weights.1 Plasmids present in the previously described strains E. coli PDK-9, R1, RP4, Sa and V5171 were used as molecular weight standards.
Pulsed-field gel electrophoresis
Intact agarose-embedded chromosomal DNA of S. dysenteriae 1 was prepared according to the procedures described earlier.9 It was digested with XbaI restriction enzyme (Gibco-BRL, Gaithersburg, MD, USA); the restriction fragments were then resolved using CHEF-DRII system apparatus (Bio-Rad Laboratories, Richmond, CA, USA) in 1% pulsed-field certified agarose in 0.5 × TBE (Tris/borate/EDTA) buffer with the following pulse times, 1–10 s for 10 h, 3–28 s for 10 h, 3–35 s for 5 h and 5–70 s for 15 h. A photograph was taken using a gel documentation system, and banding patterns were established using the criteria described previously.10 Commercially available DNA of bacteriophage λ ladder (size range, 48.5–1000 kb; Bio-Rad Laboratories) and Saccharomyces cerevisiae (size range, 225–2200 kb; Bio-Rad Laboratories) were used as the size standards.
PCR amplification of gyrA
Chromosomal DNA was prepared and purified by procedures described earlier.9 Previously recommended primers were used to amplify the QRDRs of gyrA,11 to produce a 648 bp product. Thirty μL of each reaction mixture contained 3.0 μL 10× PCR buffer (Invitrogen), 2.0 μL dNTPs (Invitrogen), 10 pmol of each primer, (Integrated DNA Technologies, Inc. USA), 1 μL of chromosomal DNA and 1 U of Taq DNA polymerase enzyme (Invitrogen). PCR conditions were adopted from elsewhere.11
Nucleotide sequencing
The PCR amplicons were purified with the GFX PCR DNA and gel band purification kit (Amersham Pharmacia, USA). They were then sequenced using the dideoxynucleotide chain termination method with an ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit (Perkin-Elmer Applied Biosystems, Foster City, CA, USA) on an automated sequencer (ABI PRISM 310) at the ICDDR, B core sequencing facility.
DNA and protein sequence analysis
The chromatogram sequencing files were inspected using Chromas 2.23 (Technelysium, Queensland, Australia), and contiguous sequences were prepared using SeqMan II (DNASTAR, Madison, WI, USA). Nucleotide and protein sequence similarity searches were performed using the National Center for Biotechnology Information (NCBI, National Institutes of Health, Bethesda, MD, USA) BLAST (Basic Local Alignment Search Tool) server on GenBank database, release 138.0.12 Multiple sequence alignments were developed using CLUSTALX 1.81.13 Sequences were manually edited in the GeneDoc version 2.6.002 alignment editor.
Nucleotide sequences accession number
The nucleotide sequences of gyrA reported in this paper were submitted to GenBank using the National Center for Biotechnology Information (NCBI, Bethesda, MD, USA) Sequin, version 5.26 under accession numbers AY692480–84 and AY695829–32.
Results and discussion
Five strains from Bangladesh and two strains from Nepal, which were isolated in 2003, were resistant to tetracycline, ampicillin, sulfamethoxazole/trimethoprim, nalidixic acid, ciprofloxacin, norfloxacin and ofloxacin. One strain from Nepal, which was isolated during 2002, was also resistant to all these drugs except for sulfamethoxazole/trimethoprim (Table 1). All eight strains were susceptible to mecillinam, ceftriaxone and azithromycin. The MIC ranges of nalidixic acid and ciprofloxacin for recent and previous epidemic S. dysenteriae 1 strains are described in the Table 1. The MIC of ciprofloxacin for these eight strains was almost the same with a value of 4–6 mg/L. Retrospective analysis of MIC of ciprofloxacin for the strains, which were collected from India during 2002, revealed that among the nine strains from India, eight had an MIC of 4–6 mg/L and the remaining one showed a very high MIC of ∼12 mg/L (Table 1).
Analysis of plasmid DNA showed that 140, 6 and 2 MDa plasmids were commonly present in all the strains regardless of origin of isolation. Based on the number and molecular mass of the plasmid, strains were categorized into four different patterns (P1–P4). Among five strains in Bangladesh, four were characterized as P1 (140, 6, 4, 2) and the remaining one was P4 (140, 52, 6, 4, 2.6, 2). In Nepal, two strains were characterized as P1 and the remaining one was P3 (140, 6, 2). Plasmid patterns P1 and P2 (140, 90–20, 6, 4, 2) were found in the previous epidemic isolates from India and Bangladesh (Table 1). The plasmid patterns of previous epidemic isolates from India and Bangladesh, recent outbreak strains in India and recent ciprofloxacin-resistant strains in Bangladesh and Nepal were almost identical except for one recent ciprofloxacin-resistant strain in Bangladesh, which had a 2.6 MDa plasmid in addition to the core plasmids of S. dysenteriae 1 (Table 1).1
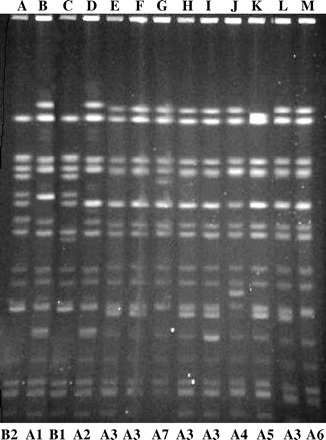
In order to scrutinize the chromosomal variation and clonal distribution of the strains from different origins and periods, we employed PFGE analysis. According to the interpretation criteria described by Tenover et al.,10 two different PFGE types designated as A and B were identified among the epidemic and sporadic strains isolated at distinct time intervals from different geographical locations (Figure 1). Type A was further subdivided into seven subtypes (subtypes A1–A7); of these, A1 and A2 were found among the strains isolated in Bangladesh and India during the epidemic of 1984. Epidemic clone A1 reemerged in Bangladesh in the following epidemic of 1994. Recent ciprofloxacin-resistant strains were grouped into subtypes A3–A7, all of which were present among the outbreak strains in India, whereas only subtype A3 was found in Bangladesh and Nepal (Table 1). On the other hand, Type B, subdivided into two subtypes (subtypes B1 and B2), was found only in Bombay during the epidemic of 1978. It appeared that ciprofloxacin-resistant S. dysenteriae 1 strains are clustered within a single PFGE type, and the existence of identical clones in Bangladesh and Nepal—and to some extent in India—are more suggestive of an epidemiologically relevant link between cases.
Generally, gyrA mutations play an important role in the development of quinolone and/or fluoroquinolones resistance in bacteria.14 In the present study, we successfully used the QRDR sequence of gyrA to investigate the clonal variation of S. dysenteriae 1 isolated in different geographical locations (India, Nepal and Bangladesh) at different time intervals. No amino acid substitution was found among the strains isolated in the epidemics of 1978 and 1984. A single amino acid substitution (Ser→Leu) at position 83 was found in the nalidixic acid-resistant strains that caused an epidemic in Bangladesh in 1994, whereas all the ciprofloxacin-resistant strains isolated in India, Nepal and Bangladesh contained a double mutation at the 83 (Ser→Leu) and 87 (Asp→Asn or Gly) positions (Table 1). Moreover, the amino acids substitutions in gyrA of the ciprofloxacin-resistant strains isolated in Bangladesh (Ser→Leu, Asp→Asn) were similar to those isolated in Nepal, but were different from the Indian strains at position 87 (Asp→Gly) (Table 1). However, strain discrimination by mutation analysis of gyrA did not correlate with the PFGE analysis, as observed in the study by Yong et al.15
Ciprofloxacin-resistant strains of S. dysenteriae 1 are found in different countries within a limited duration of time, suggesting that the strain may spread to other parts of the world very rapidly, probably traversing a similar path as previous epidemic strains. Given this knowledge, public health officials and clinicians should start preparing to forestall the spread of the epidemic.
PFGE banding patterns of XbaI-digested chromosomal DNA of representative strains of S. dysenteriae 1 collected from previous epidemics, and the recent ciprofloxacin-resistant strains of this serotype isolated from different countries. Lanes: A, PFGE type B2; B, PFGE type A1; C, PFGE type B1; D, PFGE type A2; E, PFGE type A3; F, PFGE type A3; G, PFGE type A7, H, PFGE type A3; I, PFGE type A3; J, PFGE type A4; K, PFGE type A5; L, PFGE type A3; M, PFGE type A6; respectively.
Comparative analysis of S. dysenteriae 1 strains isolated in the different epidemic periods versus recent ciprofloxacin-resistant strains of this serotype isolated from different countries
| Year of isolation . | Country of isolation . | Sporadic/ outbreak . | No. of isolate . | Plasmid patterna . | PFGE type . | MIC (mg/L) . | . | Resistance patternb . | Amino acid substitutions in gyrAc . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | CIP . | NAL . | . | Ser-83 (Wt)d . | Asp-87 (Wt)d . | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B1 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B2 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A1 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A2 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A1 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A2 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1994 | Bangladesh | outbreak | 7 | P1 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 1994 | Bangladesh | outbreak | 5 | P2 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 2002 | India | outbreak | 3 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 2 | P2 | A4 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A5 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A6 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A7 | 12 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | Nepal | sporadic | 1 | P3 | A3 | 6.0 | >256 | TETrAMPrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 4 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 1 | P4 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Nepal | sporadic | 2 | P1 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| Year of isolation . | Country of isolation . | Sporadic/ outbreak . | No. of isolate . | Plasmid patterna . | PFGE type . | MIC (mg/L) . | . | Resistance patternb . | Amino acid substitutions in gyrAc . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | CIP . | NAL . | . | Ser-83 (Wt)d . | Asp-87 (Wt)d . | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B1 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B2 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A1 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A2 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A1 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A2 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1994 | Bangladesh | outbreak | 7 | P1 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 1994 | Bangladesh | outbreak | 5 | P2 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 2002 | India | outbreak | 3 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 2 | P2 | A4 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A5 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A6 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A7 | 12 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | Nepal | sporadic | 1 | P3 | A3 | 6.0 | >256 | TETrAMPrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 4 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 1 | P4 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Nepal | sporadic | 2 | P1 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
P1, 140, 6, 4, 2; P2, 140, (90–20), 6, 4, 2; P3, 140, 6, 2; P4, 140, 52, 6, 4, 2.6, 2.
AMP, ampicillin; TET, tetracycline; SXT, sulfamethoxazole/trimethoprim; NAL, nalidixic acid; CIP, ciprofloxacin; NOR, norfloxacin; OFX, ofloxacin.
Sequence analysis of gyrA has been performed on representative strains.
Wt, wild-type; –, indicates no substitution.
Comparative analysis of S. dysenteriae 1 strains isolated in the different epidemic periods versus recent ciprofloxacin-resistant strains of this serotype isolated from different countries
| Year of isolation . | Country of isolation . | Sporadic/ outbreak . | No. of isolate . | Plasmid patterna . | PFGE type . | MIC (mg/L) . | . | Resistance patternb . | Amino acid substitutions in gyrAc . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | CIP . | NAL . | . | Ser-83 (Wt)d . | Asp-87 (Wt)d . | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B1 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B2 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A1 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A2 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A1 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A2 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1994 | Bangladesh | outbreak | 7 | P1 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 1994 | Bangladesh | outbreak | 5 | P2 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 2002 | India | outbreak | 3 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 2 | P2 | A4 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A5 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A6 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A7 | 12 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | Nepal | sporadic | 1 | P3 | A3 | 6.0 | >256 | TETrAMPrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 4 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 1 | P4 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Nepal | sporadic | 2 | P1 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| Year of isolation . | Country of isolation . | Sporadic/ outbreak . | No. of isolate . | Plasmid patterna . | PFGE type . | MIC (mg/L) . | . | Resistance patternb . | Amino acid substitutions in gyrAc . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | CIP . | NAL . | . | Ser-83 (Wt)d . | Asp-87 (Wt)d . | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B1 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1978 | Bombay, India | outbreak | 1 | P1 | B2 | 0.008 | 2 | TETrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A1 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Bangladesh | outbreak | 4 | P1 | A2 | 0.008 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A1 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1984 | Calcutta, India | outbreak | 4 | P1 | A2 | 0.012 | 2 | TETrAMPrSXTr | – | – | ||
| 1994 | Bangladesh | outbreak | 7 | P1 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 1994 | Bangladesh | outbreak | 5 | P2 | A1 | 0.047–0.25 | 64 | TETrAMPrSXTrNALr | Leu | – | ||
| 2002 | India | outbreak | 3 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 2 | P2 | A4 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A5 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P2 | A6 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | India | outbreak | 1 | P1 | A7 | 12 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Gly | ||
| 2002 | Nepal | sporadic | 1 | P3 | A3 | 6.0 | >256 | TETrAMPrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 4 | P1 | A3 | 4.0–6.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Bangladesh | sporadic | 1 | P4 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
| 2003 | Nepal | sporadic | 2 | P1 | A3 | 4.0 | >256 | TETrAMPrSXTrNALrCIPrNORrOFXr | Leu | Asn | ||
P1, 140, 6, 4, 2; P2, 140, (90–20), 6, 4, 2; P3, 140, 6, 2; P4, 140, 52, 6, 4, 2.6, 2.
AMP, ampicillin; TET, tetracycline; SXT, sulfamethoxazole/trimethoprim; NAL, nalidixic acid; CIP, ciprofloxacin; NOR, norfloxacin; OFX, ofloxacin.
Sequence analysis of gyrA has been performed on representative strains.
Wt, wild-type; –, indicates no substitution.
This study was funded by the United States Agency for International Development (USAID) under Cooperative Agreement No. HRN-A-00–96-90005-00 and ICDDR,B: Centre for Health and Population Research which is supported by countries and agencies, which share its concern for the health problems of developing countries. ICDDR,B acknowledges with gratitude the commitment of USAID and other donors to the Centre's research effort.
References
Talukder, K. A., Islam, M. A., Khajanchi, B. K. et al. (
Bennish, M. L. & Wojtyniak, B. J. (
Rahman, M. M., Huq, I., Dey, C. R. et al. (
Shahid, N. S., Rahaman, M. M., Haider, K. et al. (
Hossain, M. A., Rahman, M., Ahmed, Q. S. et al. (
Sur, D., Niyogi, S. K., Sur, S. et al. (
Datta, P. & Sen, D. (
National Committee for Clinical Laboratory Standards. (
Talukder, K. A., Islam, M. A., Dutta, D. K. et al. (
Tenover, F. C., Arbeit, R. D., Goering, R. V. et al. (
Rahman, M., Mauff, G., Levy, J. et al. (
Altschul, S. F., Gish, W., Miller, W. et al. (
Thompson, J. D., Gibson, T. J., Plewniak, F. et al. (
Lindgren, P. K., Karlsson, A. & Hughes, D. (
Author notes
1Enteric Microbiology Laboratory, Laboratory Sciences Division, ICDDR, B: Centre for Health and Population Research, GPO Box-128, Dhaka-1000, Bangladesh; 2National Public Health Laboratory, Kathmandu, Nepal; 3National Institute of Cholera and Enteric Diseases, Kolkata, India; 4Department of Bacteriology, National Institute of Infectious Diseases, Toyama 1–23–1, Shinjuku-ku, Tokyo 162, Japan