-
PDF
- Split View
-
Views
-
Cite
Cite
S. E. Hoebee, S. Angelone, D. Csencsics, K. Määttänen, R. Holderegger, Diversity of S-Alleles and Mate Availability in 3 Populations of Self-Incompatible Wild Pear (Pyrus pyraster), Journal of Heredity, Volume 103, Issue 2, March-April 2012, Pages 260–267, https://doi.org/10.1093/jhered/esr126
Close - Share Icon Share
Abstract
Small populations of self-incompatible plants may be expected to be threatened by the limitation of compatible mating partners (i.e., S-Allee effect). However, few empirical studies have explicitly tested the hypothesis of mate limitation in small populations of self-incompatible plants. To do so, we studied wild pear (Pyrus pyraster), which possesses a gametophytic self-incompatibility system. We determined the S-genotypes in complete samplings of all adult trees from 3 populations using a PCR-RFLP approach. We identified a total of 26 different S-alleles, homologous to S-alleles of other woody Rosaceae. The functionality of S-alleles and their Mendelian inheritance were verified in artificial pollination experiments and investigations of pollen tube growth. The smallest population (N = 8) harbored 9 different S-alleles and showed a mate availability of 92.9%, whereas the 2 larger populations harbored 18 and 25 S-alleles and exhibited mate availabilities of 98.4% and 99.2%, respectively. Therefore, we conclude that even small populations of gametophytic self-incompatible plants may exhibit high diversity at the S-locus and are not immediately threatened owing to reduced mate availability.
About 60% of plant species have a genetic mechanism that prevents selfing or mating with like-genotypes and are hence self-incompatible (Hiscock and McInnis 2003). The genetic mechanisms of rejection of self-pollen are well known in the case of homomorphic gametophytic or sporophytic self-incompatibility systems (Richman and Kohn 1996). In gametophytic self-incompatibility, self or foreign pollen is rejected if the S-allele of the pollen matches either of the 2 S-alleles of the receiving stigma. If the pollen donor and the pollen recipient have completely different S-genotypes, that is, share no S-allele (e.g., S1S2 × S3S4), the cross is fully compatible. If mating partners share 1 S-allele (e.g., S1S2 × S1S3), only pollen with the common allele is rejected and the cross is semicompatible. If there is surplus pollen, a semicompatible cross can still result in full fertilization, that is, indistinguishable from a fully compatible cross as based on fruit or seed set. Finally, if mating partners share both S-alleles at the self-incompatibility locus (e.g., S1S2 × S1S2), all pollen will be rejected and no fertilization occurs (De Nettancourt 2001). Under the sporophytic self-incompatibility system, pollen coat expresses proteins presenting the complete S-genotype of the pollen donor rather than its own haploid genotype. If either or both of its 2 S-alleles match either of the 2 S-alleles in a receiving stigma, the pollen tube growth and successful fertilization are prohibited and the cross is incompatible (e.g., S1S2 × S1S2 or S1S2 × S1S3). Fertilization can only occur if both pollen S-alleles differ from those expressed in the pistil, in which case the cross is compatible (e.g., S1S2 × S3S4; De Nettancourt 2001). However, sporophytic self-incompatibility relationships are often more complex than given in the simple scenario above owing to dominance effects among S-alleles (Richman and Kohn 1996; Vekemans et al. 1998).
Self-incompatibility systems in plants have gained recent interest in conservation biology and management. It has been demonstrated that a reduction in population size should lead to a reduction of genetic diversity at the S-locus in self-incompatible plants, causing reduced availability of compatible mates (Busch and Schoen 2008; Pickup and Young 2008). Hence, small populations of self-incompatible plants might exhibit reduced seed set and lowered sexual reproduction compared with larger populations as a result of mate limitation, that is, the S-Allee effect (Wagenius et al. 2007; Levin et al. 2009; Leducq et al. 2010). This S-Allee effect should be stronger in species with sporophytic than in species with gametophytic self-incompatibility systems owing to the greater restriction on mating in the former system (Vekemans et al. 1998; Busch and Schoen 2008).
The most common self-incompatibility system in plants is gametophytic self-incompatibility (De Nettancourt 2001). For this system, Wright (1939) predicted a rapid decrease in the number of S-alleles with decreasing population size. For instance, a population of 20 individuals was predicted to harbor about 6 different S-alleles, but the actual number of S-alleles depends on the rate at which new S-alleles enter a population by mutation or immigration. Wright (1939) also showed that, with increasing population size, the number of S-alleles reaches an upper plateau due to negative-frequency dependent selection at the S-locus, where rare S-alleles have a higher reproductive fitness than abundant S-alleles. Simulations of mate availability in self-incompatible plants (Vekemans et al. 1998) largely confirmed the theoretical findings of Wright (1939) and showed that mate availability should be close to 100% in a gametophytic self-incompatible population if it harbors about 10 different S-alleles and that a population of 30 individuals should have about 85% mate availability under equilibrium conditions. Based on these theoretical expectations, one would expect that small populations of gametophytic self-incompatible plants do not necessarily show strongly reduced mate availability (but see Hoebee et al. 2008), much in contrast to sporophytic self-incompatible plants where empirical studies have confirmed the occurrence of pronounced S-Allee effects (e.g., Pickup and Young 2008; Young and Pickup 2010).
Traditionally, mate limitation in populations of self-incompatible plants has been estimated by pollination experiments with pollen from a single pollen donor or mixed pollen donors (e.g., Fischer et al. 2003; Willi et al. 2005) or by using time-consuming reciprocal (diallel) crossing schemes to identify self-incompatibility genotypes across populations (Lawrence 2000). Castric and Vekemans (2004) and Busch and Schoen (2008) noticed a shortage of studies that used molecular genetic methods such as PCR (Glémin et al. 2005; Schueler et al. 2006) or reverse transcriptase (Raspé and Kohn 2002) to directly assess the number and frequency of different S-genotypes in natural populations. This, in part, reflects the few plant families for which the molecular basis of self-incompatibility has been documented.
As most woody Rosaceae (Raspé and Kohn 2007), wild pear (Pyrus pyraster (L.) Burgsd.) has a gametophytic incompatibility system (Holderegger et al. 2008). We recently applied a PCR-RFLP approach to S-genotype identification in P. pyraster and surveyed random samples from 15 populations in order to estimate S-allele diversity in this tree species. In the present paper, we give a detailed description of this PCR-RFLP approach and use it along with a complete sampling of all adult individuals in 3 populations of wild pear of different sizes (N = 8–88). We further prove the functionality of the species' self-incompatibility system in experimental pollinations and an analysis of pollen tube growth. We tested the following hypothesis: small populations of a gametophytic self-incompatible plant possess sufficiently high S-allele diversity such that mate availability is maintained at high levels.
Materials and Methods
Species and Populations Studied
The diploid wild pear, P. pyraster (synonym: P. communis ssp. pyraster (L.) A. et Gr.) is one of Europe's rarest tree species (Barengo et al. 2001). Its populations are often small (i.e., <20 adults), widely scattered, and naturally isolated. Wild pear grows in sparse, dry woodlands, at forest edges, on steep rocky slopes, or in woodland pastures (Kutzelnigg 1995; Määttänen and Holderegger 2008). Individual trees flower in late March to May showing large numbers of corymbs, each with about 20 white hermaphroditic flowers fertilized by generalist pollinators. The roundish, fleshy, and multiseeded fruits are dispersed by mammals (Kutzelnigg 1995).
We chose 3 populations of P. pyraster in Northern Switzerland that differed in census size, represented the natural range in population size, and were separated by more than 10 km: the small and very scarce population Mösli (N = 8; ∼2 individuals per hectare; 8°33′7″E/47°44′30″N, 830 m above sea level), the middle-sized and scarce population Tannbüel (N = 42; ∼5 individuals per hectare; 8°18′60″E/47°48′8″N, 475 m) and the large and dense population Effingen (N = 88; ∼73 individuals per hectare; 8°6′27″E/47°30′N, 520 m).
S-Allele Identification
We sampled leaf material from all adult individuals (N = 138) of P. pyraster in the 3 study populations. Fifty milligrams of fresh leaf material was lyophilized and ground in a mixer mill (MM 300 Retsch). DNA extraction was performed after the DNeasy 96 Plant Kit protocol (Qiagen) but using 600 μl of extraction buffer. For PCR amplification of S-alleles, we used the consensus primers FTQQYQ and anti-IIWPNV of Ishimizu et al. (1999) developed for Japanese pear (P. pyrifolia Nakai). These primers amplify fragments including the hypervariable regions and intron of the female determinant of gametophytic incompatibility, namely the S-RNase gene (Ishimizu et al. 1998; Zuccherelli et al. 2002; Charlesworth et al. 2005; Supplementary Figure S1).
PCR amplification of all samples was carried out in 20-μl reaction mixtures containing 1 ng genomic DNA, 1× PCR buffer, 1.5 mM MgCl2, 0.2 μM of each fluorescently labeled forward and reverse primer, 0.3 mM of each dNTP and 1U DNA Taq polymerase (Sigma-Aldrich). PCRs were performed on a PTC-100 thermocycler (MJ Research) with an initial step of 2 min at 94 °C, followed by 30 cycles each of 15 s at 94 °C, 15 s at 48 °C, and 2 min at 70 °C and a final step of 10 min at 72 °C. PCR products were separated on an Avant 3100 DNA Sequencer (Applied Biosystems) and scored with GENEMAPPER 3.5 (Applied Biosystems) against the internal size standard ROX500 (Applied Biosystems).
For sequencing of S-alleles and to check whether PCR fragments of identical length also had identical sequences, we selected at least 4 individuals for each identified PCR fragment. PCR products were run on 1.5% agarose gels, and fragments were excised with a gel cutter (Elchrom). To elute the PCR products, gel fragments were put into 40 μl of sterile water and stored at 4 °C for 24 h. We then performed further PCR amplifications with increased reaction volumes of 50 μl using 1-μl eluted PCR product as template. The resulting PCR products were cleaned by sodium acetate precipitation. Cycle sequencing reactions were carried out in 20 μl reaction containing BigDye Terminator (Applied Biosystems), 10 ng of cleaned PCR product, and 0.3 μM of each primer on a PTC-100 thermocycler (MJ Research) with an initial step of 20 s at 96 °C, followed by 25 cycles each of 5 s at 51 °C and 4 min at 60 °C. Sequences were separated on an Avant 3100 DNA Sequencer (Applied Biosystems), visualized with SEQUENCING ANALYSIS 3.4 and FACTURA 2.2. (Applied Biosystems), and consensus sequences were aligned in CLUSTAL X 1.81 (http://bips.u-strasbg.fr/fr/Documentation/ClustalX/). To check homology of the obtained sequences with known S-RNase sequences of other Rosaceae, a nucleotide BLAST search in GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) was performed.
Because we detected 3 S-allele fragments of identical length but with different sequences, we coupled the above PCR method with an RFLP step (i.e., the 345, 369, and 440 bp fragments were subsequently cut with Bsh1236I, PpuMI, and FspBI, respectively; Supplementary Table S1). This combined assay allowed the detection of S-alleles in P. pyraster based on their fragment length, coupled, in certain cases, with RFLPs. Restriction digests were completed at 37 °C overnight in reaction mixtures of 10 μl containing 5 μl of PCR product, 1× restriction enzyme buffer, 1 μg bovine serum albumin, and 2 U of restriction enzyme (Fermentas). Restriction patterns were visualized on 2% agarose gels against a 100 bp ladder. Note that under a gametophytic self-incompatibility system, all individuals should be heterozygous at the S-locus (Richman and Kohn 1996).
Functionality of S-Alleles
To determine the functionality of some of the identified S-alleles of P. pyraster, controlled pollinations were conducted in the 3 study populations (Sanzol and Herrero 2002). Two (Tannbüel) or 3 (Mösli and Effingen) adults were used as seed trees (mothers) and 4 trees per populations as pollen donors (fathers). Based on the known S-genotypes from PCR-RFLPs, we performed the following 6 treatments: 1) agamospermy: emasculation followed by immediate bagging (see below; N = 7 mother trees; note that the agamospermy treatments from 1 mother tree were lost during a thunder storm); 2) selfing: application of pollen from the same mother tree (N = 8); 3) incompatible cross: application of pollen from a father tree sharing both S-alleles with the mother tree (N = 2, low number because few flowering individuals had the same S-genotype; see Results); 4) semicompatible cross: application of pollen from a father tree sharing 1 S-allele with the mother tree (N = 5); 5) fully compatible cross: application of pollen from a father tree sharing no S-allele with the mother tree (N = 25); and 6) open pollination (N = 8). In all treatments, we emasculated 4 flower buds from each of 20 corymbs per mother tree prior to opening (640 flowers in total), removed all excess flowers, and bagged the corymbs with fine mesh tulle. At flower opening, we applied the same treatment to each of the 4 flowers within 3 corymbs per mother tree. Corymbs were rebagged after pollination, except for the open pollination treatment where rebagging took place 2 weeks later. Pollinations were repeated after 2 days. In autumn, we harvested the ripe fruits and determined the number of fruits per corymb as well as the number of ripe and undeveloped seeds per fruit. We then calculated the means of the relative fruit set (RFS), the seed/ovule ratio (SO), and the relative reproductive success (RRS = RFS × SO) per treatment per mother plant.
The agamospermy treatment produced no fruit and was thus excluded from statistical analysis (see Results). The other 5 treatments were grouped into 3 classes, that is, incompatible (selfing and incompatible crosses; N = 9), compatible (semicompatible and fully compatible crosses; N = 25), and open pollination (N = 8). Explorative analysis showed that there was neither an effect of population nor of the interaction between population and the 3 above classes. We therefore performed one-way ANOVAs on RFS, SO, and RRS in SPSS 11.0 (SPSS). Orthogonal linear contrasts (not assuming equal variances) tested for differences 1) of incompatible versus compatible and open-pollinated treatments and 2) of compatible versus open-pollinated treatments. Residuals showed no deviation from normal distribution.
We also assessed the functionality of S-alleles by analyzing pollen tube growth. We again emasculated 3 to 6 flower buds per corymb of 4 mother trees in the Mösli population and rebagged them. After 2 days, the following pollinations were carried out with surplus pollen on all of the flowers per corymb: 1) fully compatible crosses (N = 4; 89 styles studied), 2) semicompatible crosses (N = 2; 51 styles), 3) incompatible cross (N = 1; 25 styles), and 4) selfing (N = 2; 44 styles). After 8 days, the flowers were fixed in FAA (formaldehyde–acidic acid–ethanol 1:1:18) and stored at 4 °C. Flowers were then washed 3 times with distilled water, soaked with Na2SO3 overnight and autoclaved at 110 °C for 10 min. Styles and gynoecia were excised, stained in 0.1% aniline blue and 0.1 N K3PO4 (Linskens and Esser 1957) and immediately screened under UV-epifluorescence using an Olympus microscope. Pollen tubes showed blue-green fluorescence. Because incompatible pollen tubes either do not germinate or are stopped just underneath the stigma surface in Pyrus (Sanzol and Herrero 2002; Sanzol et al. 2003), we could easily screen whether pollen tubes had penetrated style tissue toward the ovaries as expected in semi- or fully compatible crosses.
To confirm the inheritance of S-alleles, we determined the S-genotypes in a sample of 26 progenies from the above controlled pollinations (123 offspring in total). DNA extraction of embryos, which were carefully dissected from seeds and separated from all maternal tissue, followed the protocol given above. We determined the S-genotypes of all mothers, fathers and offspring using the PCR-RFLP approach described above. We then checked the correctness of the performed pollinations by applying 4 nuclear microsatellites developed for Japanese pear (P. pyrifolia; NH007, NH009, NH011, and NH013; Yamamoto et al. 2002), already used for P. pyraster by Holderegger et al. (2008). Microsatellite amplification was performed in 20 μl reaction mixtures containing 1 ng genomic DNA, 1× PCR buffer, 1.5 mM MgCl2, 0.2 μM of each forward and reverse primer, 0.2 mM of each dNTP and 0.2 U of Taq polymerase (Sigma-Aldrich). PCRs were performed on a PTC-100 thermocycler (MJ Research) with an initial step of 2 min at 94 °C, followed by 35 cycles each of 60 s at 94 °C, 60 s at 55 °C (58–52 °C touchdown for NH013) and 2 min at 70 °C and a final step of 30 min at 72 °C. PCR products were separated on Spreadex Ready-to-Use Mini Gels (Elchrom) against M3 DNA ladder (Elchrom) and visualized by SYBR-gold staining. The size of the amplified fragments was determined using LABWORKS 4.0 (UVP).
S-Allele Diversity in 3 Natural Populations
For the 3 populations studied, we determined the S-allele diversity per population (without null alleles; see Results) and tested the frequencies of S-alleles within each population for isoplethy (i.e., uniform distribution of S-alleles; Wright 1939) using simulated equal-frequency populations for comparison. We also tested whether the 3 populations showed different S-allele frequency distributions. For this aim, we used the Markov chain Fisher's exact test implemented in CHIFISH with 500 000 iterations (Ryman 2006). Finally, we identified the percentage of adult trees with unique S-genotypes per population as well as the mean percentage of compatible trees per population (i.e., mate availability).
Results
S-Allele Identification
We identified a total of 26 different S-alleles in the 3 populations studied. These S-alleles were highly variable and had fragment lengths between 334 and 2000 bp. We obtained only partial sequences for fragments longer than 550 bp (Supplementary Figure S1; only complete sequences were submitted to GenBank: accession numbers DQ394811 to DQ394831). A nucleotide BLAST search showed substantial sequence similarity (up to 99%) with S-RNase sequences of other woody Rosaceae such as cultivated Pyrus spp. (e.g., Norioka et al. 1996; Ishimizu et al. 1998; Takasaki et al. 2006) and Malus spp. (Janssens et al. 1995; Van Nerum et al. 2001), as well as wild Sorbus aucuparia and Craetagus monogyna (Raspé and Kohn 2002). We also detected 3 S-allele fragments of identical length but different sequences. In these cases, additional RFLPs were applied (Supplementary Table S1; data deposited in the Dryad repository: doi: 10.5061/dryad.hd8gs614).
We observed a high frequency of null alleles (17.8%), most likely due to a lack of complementarity of the universal primers used, that is, for some individuals only a single S-allele could be amplified. Because we were always able to amplify at least 1 S-allele for each individual, we assumed that a single null S-allele occurred in each of the studied populations. Consequently, we treated S-genotypes having the same S-allele and a null S-allele (e.g., S1null and S1null) as identical S-genotypes, although they could equally well have constituted 2 different S-genotypes if more than a single null allele was present. However, the benefit to this assumption of a single null S-allele per population was that we achieved a maximum estimate of mate limitation within populations (see below).
Functionality of S-Alleles
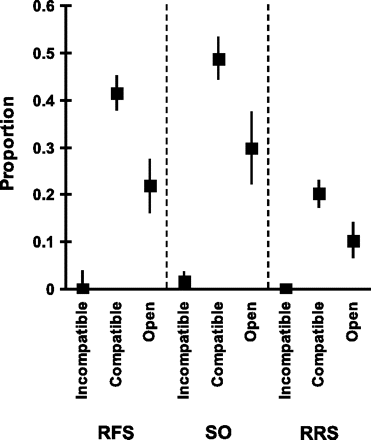
No fruit was produced in the agamospermy treatment. In contrast, both fruit and seed set were achieved after incompatible, compatible, and open pollination treatments (Figure 1). RFS in incompatible pollinations was low with a mean of 0.028 ± 0.020 (standard error). It was higher in open (0.099 ± 0.046) and compatible pollinations (0.412 ± 0.034), and there was a significant difference among treatments (F2,39 = 29.553, P ≤ 0.001). Incompatible pollinations also showed the lowest value of SO (0.064 ± 0.045), whereas open (0.167 ± 0.083) and compatible (0.486 ± 0.219) pollinations had significantly higher SO-values (F2,39 = 16.927, P ≤ 0.001). The same significant ranking order was found for RRS with means of 0.007 ± 0.005, 0.023 ± 0.012 and 0.200 ± 0.027 in incompatible, open and compatible pollinations, respectively (F2,39 = 14.976, P ≤ 0.001). Incompatible pollinations were different from both open and compatible pollinations, and open and compatible pollinations were different from each other for RFS, SO, and RRS (P ≤ 0.006 in all cases; Figure 1). Note that the RRS of compatible crosses was about 20%, whereas that of open pollinations was considerably lower with only 2%, a result potentially indicating pollen limitation in the natural populations of P. pyraster studied.
RFS, SO, and RRS in incompatible (selfing and incompatible crosses), compatible (semicompatible and fully compatible crosses), and open pollination in artificial pollinations of wild pear trees of known S-genotypes.
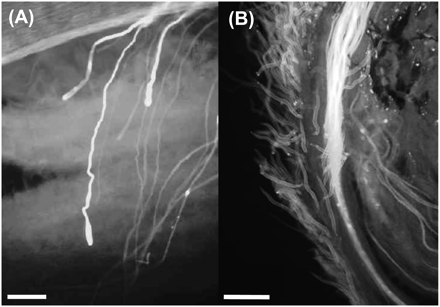
Analysis of pollen tube growth showed that pollen tubes in incompatible and selfing treatments only grew to just below the stigma surface (Figure 2A). In contrast, pollen tubes from both semicompatible and fully compatible crosses reached the ovaries (Figure 2B).
Fluorescence microscopy of pollen tube growth. (A) Pollen tubes from an incompatible cross between wild pear parental trees having the same S-genotype. Pollen tubes are stopped (as indicated by swollen tips) just underneath the surface of the stigma. (B) Pollen tubes from a compatible cross between wild pear parental trees having different S-genotypes grow down the style toward the ovaries (bar = 100 μm in both cases).
Subsequent genetic testing using 4 microsatellite loci showed that 13 out of the 123 analyzed offspring (10.6%) originated from pollen contamination. Hence, the majority of offspring resulted from the experimental pollinations. These contaminations also included 3 offspring from 2 selfing treatments (diminishing the observed seed set of selfing as given in Figure 1). However, we could verify that selfing resulted in 3 offspring. Furthermore, S-alleles showed Mendelian inheritance, and open-pollinated progenies only contained S-alleles already identified in the adult trees.
S-Allele Diversity in 3 Natural Populations of Different Size
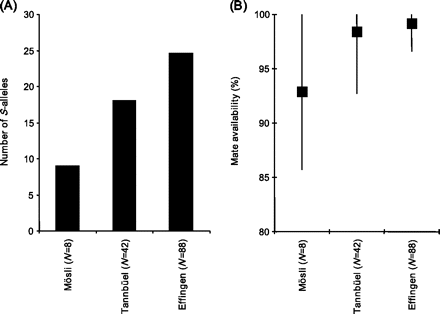
The smallest population, Mösli, harbored 9 different S-alleles (Figure 3), and 75% of the adults had unique S-genotypes. The Tannbüel population had 18 different S-alleles and 76% trees with unique S-genotypes, and the largest population, Effingen, had 25 S-alleles and 73% trees with unique S-genotypes. The frequencies of S-alleles within the 3 populations did not significantly differ from isoplethy (Mösli: P = 0.993; Tannbüel: P = 0.814; Effingen: P = 0.054), but frequencies significantly differed among the 3 populations (P ≤ 0.001; Supplementary Figure S2 ). Mate availability was high with a mean of 92.9% in the small population (Mösli), whereas the middle-sized population (Tannbüel) and the large population (Effingen) had mean mate availabilities of 98.4% and 99.15%, respectively (Figure 3).
Number of S-alleles (A) and mean mate availability and its range across individuals (B) in 3 populations of wild pear differing in census size (N).
Discussion
S-Alleles Diversity in Wild Pear
In 3 fully sampled populations of wild pear with a gametophytic self-incompatibility system, we detected a total of 26 alleles at the S-RNase locus. Given the total of 138 adult trees sampled, this represents a higher than expected S-allele diversity based on theoretical expectations for gametophytic self-incompatibility systems. Comparably high estimates of S-allele diversity are theoretically predicted for populations of about 1000 individuals (Wright 1939, model with mutation rate 10−3; Vekemans et al. 1998). However, the total number of S-alleles found in the present study agreed with empirical data on S-allele diversity in natural populations of other species. Lawrence (2000) reviewed the existing literature on empirical studies of S-allele diversity. The reviewed studies had all used the traditional diallel crossing approach to define the number of S-alleles. Lawrence (2000) showed that populations exhibiting gametophytic self-incompatibility harbor between 15 and 41 different S-alleles and that multiple populations of the same species can have up to 45 different S-alleles. In contrast to Lawrence (2000), Castric and Vekemans (2004) reviewed studies that used molecular genetic methods to estimate S-allele diversity. These authors found that populations of species with a gametophytic self-incompatible system possess between 10 and 36 different S-alleles, although the maximum number of individuals tested in any of these studies was only 67. When looking specifically at molecular genetic studies of gametophytic self-incompatibility in woody Rosaceae, it is evident that our estimate of 26 different S-alleles in 3 populations of wild pear was not unusually high. Raspé and Kohn (2002) detected 17 S-alleles in 13 individuals of Crataegus monogyna and Raspé and Kohn (2007) found 30 S-alleles in 2 populations each with 20 individuals sampled for Sorbus aucuparia. A slightly lower value of 15 S-alleles was found by Schueler et al. (2006) in a completely sampled population of 168 wild cherry trees (Prunus avium). About 20 different S-alleles were found in cultivated European pear (P. communis L.; Goldway et al. 2009; Sanzol 2009).
The S-allele fragments detected in wild pear varied between 334 and 2000 bp in length. This range was similar to that found in Japanese pear (P. pyrifolia) with S-allele lengths between 352 and 1347 bp and also in cultivated pear (P. communis) with S-allele lengths between 641 and 2217 bp (Sanzol 2009; note that the same consensus primers had been used for all these Pyrus species; Takasaki et al. 2006). The fragments in wild pear were highly variable and difficult to align (Supplementary Figure S1). Nevertheless, they showed high similarities to S-allele sequences of other Rosaceae, consistent with the shared history of gametophytic self-incompatibility in woody Rosaceae (Vieira and Charlesworth 2002).
Functionality of S-Alleles and Mate Availability in Wild Pear
We found no evidence for agamospermy in P. pyraster. Furthermore, seed set and the patterns of pollen tube growth, after controlled pollinations of trees with known S-genotypes, clearly showed that the species possesses a functional homomorphic multiallelic and gametophytic self-incompatibility system. Fruit set, seed set, and reproductive success were all much lower in self-pollinated and incompatible crosses than in semicompatible or fully compatible crosses and open pollination (Figure 1). Similarly, pollen tubes of incompatible (including self) pollinations never grew further than just beneath the stigma surface (Figure 2A).
Our largest population, with a census size of 88 adult trees, harbored 25 S-alleles. This contrasts with Wright's (1939) balanced model, which only predicts 25 different S-alleles in populations of about 100 individuals under an unlikely high mutation rate of 10°. Under a more reasonable mutation rate of <10−3, only populations of more than 1000 individuals should exhibit such a high number of S-allele, a prediction largely in congruence with Vekemans et al. (1998). In our smallest population (Mösli; N = 8), we identified 9 S-alleles. In accordance with our results, Lawrence (2000) stressed that the number of S-alleles found within populations is usually greater than that predicted by theory. A possible explanation for the comparable high S-allele diversity found in our 3 populations could be a high immigration rate of new S-alleles, which would subsequently experience positive frequency dependent selection (Castric and Vekemans 2004). However, our 2 larger populations, Tannbüel and Effingen (N = 42 and 88, respectively), only shared about 65% of their alleles, and the open-pollinated progenies in our artificial pollination experiments did not reveal any novel S-alleles. This suggested limited S-allele immigration rates. Alternatively, we cannot exclude that gene flow from cultivated to wild pear occurred (Kutzelnigg 1995), as cultivars of P. communis were also grown in the regions of our 3 study populations. Such gene flow could potentially have increased S-allele diversity in populations of wild pear. However, there are no studies available yet that explicitly tested for the amount of gene flow between cultivated and wild pear.
The high number of different S-genotypes we detected in wild pear translated into high mate availabilities, between 92.9% for the smallest population (Mösli) and 99.15% for the largest population (Effingen; Figure 3). Therefore, mate availability was not severely limited even in a very small population of wild pear. This result was in good agreement with theoretical expectations on mate availability in small populations of plants with a gametophytic self-incompatibility system, which demonstrate that only populations harboring less than 5 different S-alleles should show reduced mate availabilities below 90% (Vekemans et al. 1998).
Conclusions
Our original hypothesis was that small populations of gametophytic self-incompatible plants can possess sufficiently high numbers of different S-alleles such that the availability of compatible mates is maintained at high levels. We demonstrated the validity of this hypothesis for populations of wild pear, which exhibited mate availabilities above 90%, even in 1 population that harbored only 8 adult trees. Thus, we did not find a significant S-Allee effect in P. pyraster. In contrast to plants with a gametophytic self-incompatibility system such as P. pyraster, S-Allee effects clearly occur in small populations of plant species having sporophytic self-incompatibility systems (Vekemans et al. 1998). When dealing with the management of small plant populations in nature conservation, we suggest distinguishing between species with sporophytic and gametophytic self-incompatibility systems. The former are potentially threatened by small population size and S-Allee effects, whereas the later might be much less affected by mate limitation under small population size.
Funding
Swiss Agency for the Environment BAFU (project “Protection and Use of Forest Genetic Resources”).
We thank Felix Gugerli for help with artificial pollinations, the private owners as well as the forest and conservation authorities for the permission to work on their land, and Ed Newbigin and 3 anonymous referees for comments on the manuscript.
References
Author notes
Corresponding Editor: John Stommel