-
PDF
- Split View
-
Views
-
Cite
Cite
J. A. da Costa-Nunes, A. M. Bhatt, S. O'Shea, C. E. West, C. M. Bray, U. Grossniklaus, H. G. Dickinson, Characterization of the three Arabidopsis thaliana RAD21 cohesins reveals differential responses to ionizing radiation, Journal of Experimental Botany, Volume 57, Issue 4, March 2006, Pages 971–983, https://doi.org/10.1093/jxb/erj083
Close - Share Icon Share
Abstract
The RAD21/REC8 gene family has been implicated in sister chromatid cohesion and DNA repair in several organisms. Unlike most eukaryotes, Arabidopsis thaliana has three RAD21 gene homologues, and their cloning and characterization are reported here. All three genes, AtRAD21.1, AtRAD21.2, and AtRAD21.3, are expressed in tissues rich in cells undergoing cell division, and AtRAD21.3 shows the highest relative level of expression. An increase in steady-state levels of AtRAD21.1 transcript was also observed, specifically after the induction of DNA damage. Phenotypic analysis of the atrad21.1 and atrad21.3 mutants revealed that neither of the single mutants was lethal, probably due to the redundancy in function of the AtRAD21 genes. However, AtRAD21.1 plays a critical role in recovery from DNA damage during seed imbibition, prior to germination, as atrad21.1 mutant seeds are hypersensitive to radiation damage.
This paper is available online free of all access charges (see http://jxb.oxfordjournals.org/open_access.html for further details)
Introduction
The maintenance of genome integrity is dependent on accurate chromosome segregation and repair of DNA damage during the cell cycle. Both processes are essential for the faithful transmission of the genetic information to daughter cells and for the survival of the organism. In yeast (Schizosaccharomyces pombe and Saccharomyces cerevisiae) and Caenorhabditis elegans, RAD21/SCC1/MCD1/CHO-2 (from now on referred as RAD21) is one of the genes involved in these essential functions since it has been implicated not only in DNA double-strand break (DSB) repair (Birkenbihl and Subramani, 1992; Boulton et al., 2002) but also in sister chromatid cohesion in mitotic cells (Guacci et al., 1997; Michaelis et al., 1997; Chan et al., 2003; Mito et al., 2003). REC8, the meiotic homologue of RAD21 has also been found to be essential for chromosome cohesion and DNA DSB repair in meiotic cells (Molnar et al., 1995; Klein et al., 1999; Watanabe and Nurse, 1999). Genes belonging to the RAD21/REC8 gene family have been identified in several organisms, from yeast to humans (Birkenbihl and Subramani, 1992; Molnar et al., 1995; McKay et al., 1996; Guacci et al., 1997; Michaelis et al., 1997; Heo et al., 1998; Losada et al., 1998; Bhatt et al., 1999; Klein et al., 1999; Watanabe and Nurse, 1999; Warren et al., 2000; Dong et al., 2001; Pasierbek et al., 2001) supporting their central role in cell division. Unlike most organisms, which have only one mitotic RAD21 gene, Caenorhabditis elegans (C. elegans) (Pasierbek et al., 2001), Arabidopsis thaliana (Arabidopsis; Dong et al., 2001; this study), and rice (Oryza spp.) possess three different RAD21-like genes. By contrast, both Arabidopsis (Bai et al., 1999; Bhatt et al., 1999; Mercier et al., 2003) and C. elegans (Pasierbek et al., 2001) have only one REC8 gene important for both sister chromatid cohesion and DNA DSB repair during meiosis.
In several organisms, the RAD21 gene is transcribed and translated in a cell cycle-regulated fashion since sister chromatid cohesion has to be established when RAD21 (as part of the cohesion complex) is loaded on to the nascent sister chromatids during S-phase with the assistance of other proteins; ultimately, the loading of this cohesin complex maintains cohesion between the two sister chromatids (Guacci et al., 1997; Michaelis et al., 1997; Uhlmann and Nasmyth, 1998; Skibbens et al., 1999; Tóth et al., 1999; Ciosk et al., 2000; Mayer et al., 2001). In budding yeast, Xenopus, and humans this complex has been shown to contain RAD21, and three other proteins (SMC1, SMC3, and SCC3) (Guacci et al., 1997; Michaelis et al., 1997; Losada et al., 1998, 2000; Tóth et al., 1999; Sumara et al., 2000). In fission yeast the cohesin complex lacks SCC3 (Tomonaga et al., 2000).
The cohesion between sister chromatids is maintained until the metaphase-to-anaphase transition, when serine residues at the RAD21 cleavage sites are phosphorylated (Alexandru et al., 2001) and targeted for proteolytic cleavage by the separase ESP1 (Ciosk et al., 1998; Uhlmann et al., 1999, 2000; Waizenegger et al., 2000; Hauf et al., 2001). The cleavage releases the ties between the two sister chromatids allowing them to be separated to opposite directions by the mitotic spindle.
In addition to their function in chromosome segregation RAD21, and its meiotic homologue REC8, have also been implicated in DNA double-strand break (DSB) repair (Birkenbihl and Subramani, 1992; Klein et al., 1999). The fission yeast rad21-45 mutant is viable under normal growth conditions but is hypersensitive to DNA damage (Birkenbihl and Subramani, 1992), similarly budding yeast RAD21 mutants are also sensitive to gamma rays (Heo et al., 1998). Finally, the depletion of cohesins from Xenopus cells and RAD21 from chicken cells also results in the persistence of chromosomal breaks in gamma ray irradiated cells (Losada et al., 1998; Sonoda et al., 2001), thus suggesting a role for RAD21 in DNA DSB repair after DSB induction. Further support for the role of RAD21 in DNA DSB repair comes from the observation that in yeast cells RAD21 is specifically recruited to the DNA damaged sites after induction of DSB in G2 (Ström et al., 2004; Ünal et al., 2004). Previously, Kim et al. (2002) had also shown that, in human cells, SMC1 (a component of the cohesin complex) is also recruited by DNA repair proteins to the DNA DSB damaged sites.
It is currently thought that the RAD21 DNA DSB repair activity is due to its cohesin function ensuring correct chromosome topology and physical proximity of the sister chromatids (in the S and G2 phases), hence facilitating DNA repair, rather than to a catalytic function of RAD21 in DNA repair. This is supported by data from Sjögren and Nasmyth (2001) showing that RAD21 per se attached to the DNA of non-joined sister chromatids is not enough to repair DNA DSB. Despite these associations between RAD21 and DNA repair, very few reports have been published on how RAD21 expression responds to DNA damaging agents. According to McKay et al. (1996) expression of the human RAD21 gene is not enhanced by exposure of diploid fibroblasts to DNA damaging agents like gamma radiation.
In this study, the cloning of three different RAD21 cDNAs from Arabidopsis, their expression profile and differential response to DNA damage are described as well as reporting the phenotype of the atrad21.1 mutant after exposure to ionizing radiation.
Materials and methods
Plant material
The ap1-1/cal-1 double mutant (Landsberg erecta) and T-DNA lines salk_044851 and salk_076116 (Alonso et al., 2003) were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/). Seedlings were grown on germination medium plates (GM; Gibco-BRL) in a growth cabinet (Sanyo Gallenkamp PLC Fi-Totron TC) with a light cycle of 16 h at 22.5 °C alternating with 8 h of darkness at 19.5 °C; seedling tissue was collected 7 d after germination. For the remaining samples, tissue was collected from plants grown on soil (1:1 v:v mixture of compost and vermiculite; Levington Universal Extra Compost, Levington, UK, and Silvaperl Vermiperl medium grade, respectively) and grown in a Sanyo Gallenkamp PLC growth cabinet in 16 h of light at 20 °C alternating with 8 h of darkness at 15 °C. Plant transformation was carried out by dipping inflorescences (Clough and Bent, 1998) in Agrobacterium tumefaciens GV3101 containing pMP90RK. Plant transformants were selected on GM medium containing 30 μg ml−1 hygromycin.
DNA damage assays (ionizing radiation and UV)
Arabidopsis plants at 23-d-old were irradiated with 30 krad (300 Gy at a rate of 2.6 Gy min−1) of gamma rays using 137Ce (RX30 55M Irradiation, Graviton Industries Lda.). Arabidopsis imbibed and stratified seeds and 5-d-old seedlings grown on GM medium were irradiated with 0, 50, 100, and 150 Gy (3.6 Gy min−1) of X-rays using tungsten (Müller MG 150, Philips PW 2184/00, Monitor SN4). Aerial tissues were collected 1 h after irradiation and used for RNA extraction as described in Doutriaux et al. (1998). UV-B irradiation of 3–4-week-old plants was carried out as in West et al. (2000). Hypersensitivity of plants to bleomycin was assayed as described in West et al. (2002).
RNA extraction
Total RNA was extracted using guanidine hydrochloride (Logemann et al., 1987), Trizol (Gibco-BRL) or a RNeasy Plant Qiagen total RNA extraction kit. Poly(A) RNA was purified from inflorescences using the PolyATract® mRNA Isolation System IV (Promega).
Cloning of AtRAD21 cDNAs
General molecular biology protocols were used as described in Sambrook et al. (1989). Primers for amplifying and sequencing AtRAD21 cDNAs were based on the gene predictions obtained from the Arabidopsis sequencing consortium, Genscan (http://bioweb.pasteur.fr/seqanal/interfaces/genscan.html) and NetGene2 programs (http://genome.cbs.dtu.dk). Total RNA was treated with RNase free DNase I (Gibco-BRL) and 1 μg of total RNA was used for cDNA synthesis as described in the manufacturer's protocols (AMV Reverse transcription: Promega; Superscript™ II Gibco-BRL). Full-length cDNAs for AtRAD21.1, AtRAD21.2, and AtRAD21.3 were amplified (35 cycles) with Taq/Pwo using primers CR1 (5′-ATGTTTTACTCGCATTGTCTAG) and CR10 (5′-GACACATATACTTTAGATCGTC) for AtRAD21.1, primers ACO5 (5′-ATGTTTTATTCACATACGCTTTTGG) and ACO3 (5′-TCACGTTTGAACCTTAGAAAAAAG) for AtRAD21.2, and primers AtRAD21-3-5 (5′-AGGAATGTTTTATTCGCAGTTTATA) and ATRAD21-3-3 (5′-ACCAAACTAGCTATCTGCAACTATATG) for AtRAD21.3, respectively. An aliquot of each PCR reaction was reamplified, using the same primers, to increase the amount of PCR product, which were subsequently cloned in the pGEM-T Easy (Promega) vector and sequenced. Sequence data was assembled from overlapping PCR products, spanning the entire length of the cDNA. PCR products used to sequence the junction between the 9th intron and the 10th exon in the two variants of AtRAD21.1 were amplified with 35 PCR cycles using primers CR11 (5′-CAAGCTTTTTGTGGTCTGGA) and CR22 (5′-CCACAGGAACCCCTAAAGATA). The products of the AtRAD21.1 splice variants were cloned in the pGEM-T Easy (Promega) vector, and sequenced.
5′-RACE and 3′-RACE products
cDNA synthesis for 5′-RACE and 3′-RACE was carried out according to the manufacturer's instructions (GIBCO-BRL 5′-RACE kit version 2.0 or 5′/3′-RACE Kit Roche-Boehringer Mannheim). 5′-RACE of AtRAD21.1 was performed using the Abridged Anchor Primer (5′-GGCCACGCGTCGACTAGTACGGGIIGGGII GGGIIG; GIBCO-BRL) and 5HOM1 (5′-GCTGCATCTCTTCGCAACTTG). A second PCR was performed using Abridged Universal Amplification Primer primer (AUAP; 5′-GGCCACGCGTCGACTAGTAC) and 5HOM2 (5′-CTCTTCCATGTCAAACCTTTCC). 3′-RACE of AtRAD21.1 was obtained with the Adaptor primer (5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTT) and 3HOM1 (5′-GGAAAGGTTTGACATGGAA), followed by a second PCR with 3HOM2 (5′-CATGAGACTTTCTCTACCA) and the same adaptor primer. 5′-RACE of AtRAD21.2 was carried out with the oligo d(T) anchor primer (5′-GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTV) and 5CO-1.5 primer (5′-CCAAGTCGAACTCGTCCAGAT). The PCR products were used for a second PCR using the adaptor primer (5′-GACCACGCGTATCGATGTCGAC) and 5CO-2 (5′-GCTTGAGGCAAAGTAACAGAC). A third PCR reaction was required with the 5CO-4 primer (5′-GGAAACATTATATTATCAACAGTG) and the adaptor primer. 3′-RACE of AtRAD21.2 was performed as follows, cDNA was synthesized using oligo-dT Anchor primer (5′-GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTV), subsequently 3′-RACE cDNA was amplified using the adaptor primer (5′-GACCACGCGTATCGATGTCGAC) and 3CO-1 (5′-CTGGAGGTAGGTGGCAATTC), the amplification products were subjected to a second PCR with 3CO-2 (5′-GGGAGAGCAAGAGCCTTGG) and the adaptor primer (5′-GACCACGCGTATCGATGTCGAC). These products were cloned in the pGEM-T Easy (Promega) vector, and sequenced. Similarly, the 5′ and 3′-UTR of AtRAD21.3 were amplified using the following gene-specific primers 5AT3-1 (5′-GGCACCTCAACTAATCCAGG), 5AT3-2 (5′-GCAAGATCTTCAACCTGCTCT) and 5AT3-3 (5′-GCGTTGTGATCAATACCTGGA) for 5′-RACE, and 3AT3-1 (5′-GGGAGACAATGATGAAATGGATA) and 3AT3-2 (5′-GCACATGACACAGGATTTTTGA) for 3′-RACE.
Predicted protein analysis
Protein sequence prediction and analysis was carried out with the Translate tool from Expasy (http://www.expasy.ch/tools/dna.html), PSORT (http://psort.nibb.ac.jp/), PROSITE (http://www.expasy.ch//tools/scanprosite), and PESTfind (https://emb1.bcc.univie.ac.at/content/view/21/45). Protein alignment was carried out with the program ClustalX1.8 (Person et al., 1997). Phylogenetic trees were constructed with the program Phylip 3.5 (Felsenstein, 1993).
Northern analysis
Total RNA was subjected to electrophoresis on a 1.2% formaldehyde agarose gel, blotted onto Hybond-NX membrane (Amersham Pharmacia) and crosslinked by UV at 70 000 μjoules cm−2. cDNA probes (PCR products) specific for the AtRAD21.1, AtRAD21.2, AtRAD21.3, AtRAD51, and β-tubulin gene family were radiolabelled with α-32P dCTP by random priming, and RNA filters were sequentially hybridized with AtRAD21.1, AtRAD21.3, and, finally, β-tubulin probe, with AtRAD51 followed by the β-tubulin probe, or AtRAD21.2 and the β-tubulin probe. All filters were washed at high stringency and hybridization signals were detected by autoradiography with Kodak X-Omat X-ray film (Kodak).
Knock-out lines genotyping
T-DNA SALK line salk_044851 was genotyped using the primers LBa1 (5′-TGGTTCACGTAGTGGGCCATCG) (Alonso et al., 2003) and 51L (5′-GAGATGGTCACACAGAGAATTTAG), RBa1 (5′-GTGCAATCCATCTTGTTCAATCATG) and 51R (5′-CTCCTCTCAGGACAGTCAGTATG). The T-DNA line salk_076116 was genotyped using the primers LBa1 and 16L (5′-CTGTGTCATGTGCATTTTCCATGG), RB1 (5′-ATTAGGCACCCCAGGCTTTACACTTTATG) (McElver et al., 2001), and 16R (5′-CCGTGTAGAGGATTTACAGGTTG). Plant DNA was extracted using the Edwards et al. (1991) method.
Expression analysis by semi-quantitative RT-PCR
1 μg of total RNA (DNase I-treated) was used for the cDNA synthesis using gene-specific primers for each AtRAD21 gene. 10 μl of the (1:10 dilution of the) cDNA synthesis reaction was added to the 40 μl of the PCR mix and the PCR reaction carried out for 25 amplification cycles (20 cycles for the β-tubulin gene family amplification). AtRAD21-1 498 bp PCR product amplified with primers: CR1 and 5HOM2; AtRAD21.2 530 bp PCR product amplified with primers: ACO5 and 5-CO6 (5′-GGTTCTGTTGATTGGTCGAC); AtRAD21.3 701 bp PCR product amplified with primers: ATRAD21-3–5 and AT5 (5′-GATCTTCAACCTGCTCTTC); β-tubulin-specific PCR products were amplified as controls with primers: TUB-R (5′-GTGAACTCCATCTCGTCCAT); TUB-L (5′-CCTGATAACTTCGTCTTTGG) (Knight et al., 1999). 30 μl of each PCR reaction was used for Southern analysis, and the hybridization signals were analysed using a Phosphorimager (Bio-Rad). For each tissue, four independent total RNA samples were used to synthesize the four independent cDNA synthesis reactions (RT-PCR analysis). In addition to the Phosphorimager analysis, autoradiography was also performed using Kodak X-Omat×ray film (Kodak).
RT-PCR analysis of knock-out mutants
Expression analysis of the mutants was carried out with the following primers for T-DNA line salk_044851; upstream of T-DNA insertion: CR1 and CR4 (5′-TCTAGAGTACTCATCTCAGG); downstream of T-DNA insertion: CR11 and CR22. For the T-DNA line salk_076116 the following primers were used; Upstream of T-DNA insertion: ATRAD21-3-5 and ATUP (5′-GGCTTCAGAGACTCCTTCCATCG); downstream of T-DNA insertion: ATRAD21-3-3 and ATDW (5′-CGTCTTCTAGAAAACAGTGGATGG). TUB-L and TUB-R primers were also used. 4 μl of the (1:1 dilution of the) cDNA synthesis reaction was added to the 21 μl of the PCR mix and the PCR reaction carried out for 28 amplification cycles (30 cycles for the AtRAD21.1 gene).
Southern analysis
Agarose gels were blotted to Hybond NX (Amersham Pharmacia) and DNA was crosslinked with UV at 70 000 μjoules cm−2, as described in Sambrook et al. (1989). Southern blot membranes were hybridized with the radiolabelled AtRAD21 gene-specific probes and the β-tubulin probe. Hybridization and washes were carried out at high stringency.
DIF1 promoter–uidA fusion
A 535 nucleotide PCR product corresponding to the 520 nucleotides of the DIF1 promoter and codons for the first five amino acids of DIF1/SYN1 was cloned upstream of the uidA gene, replacing the CaMV 35S promoter of pSLJ4D4 (Jones et al., 1992) to generate a DIF1–uidA translational fusion. The PCR amplification introduced a NcoI site, and also altered an alternative splice site identified in DIF1/SYN1 (Bai et al., 1999). The promoter–uidA fusion was subcloned into the binary vector pVKH18 (Batoko et al., 2000) and introduced into Arabidopsis plants by the inflorescence dip method (Clough and Bent, 1998). 26 independent transformants were analysed for uidA expression as described in Jefferson et al. (1987) and Rodrigues-Pousada et al. (1993).
Results
Arabidopsis has three AtRAD21 genes
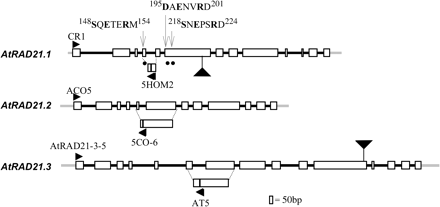
Three genes homologous to REC8/RAD21 were identified by BLAST searches of the Arabidopsis thaliana genome sequence (http://www.kazusa.or.jp/kaos/ and http://www.mips.gsf.de/thal/db) (The Arabidopsis Genome Initiative, 2000) queried with the Arabidopsis REC8 homologue. Two of the RAD21 homologues are on chromosome V, AtRAD21.1 (At5g40840) and AtRAD21.3 (At5g16270), while the third, AtRAD21.2 (At3g59550) is on chromosome III. The genomic sequences of AtRAD21.1, AtRAD21.2, and AtRAD21.3 cDNA span the nucleotides 34060–38168 from the P1 clone MHK7 (accession number AB011477), 20212–23446 of the BAC T16L24 clone (accession number AL138659), and 60806–66353 of BAC clone T21H19 (accession number AL391148), respectively.
The predicted sequence of AtRAD21.1 (wild type, Col-0) was confirmed by cloning and sequencing the full-length cDNAs, and two variants were identified which differ by a single amino acid due to an extra codon at the junction of the 9th intron and the 10th exon (variant 1; accession number AF400126). The extra codon inserts a glutamine at position 716 (716Q) in the longer AtRAD21.1 variant. Both transcripts are represented equally in both open and unopened flower RNA as approximately 50% of the clones sequenced are of each class. The adjacent 5′-untranslated region (5′-UTR) and 3’-UTR sequences were also cloned and sequenced (70 bp and 184 bp, respectively). Both AtRAD21.1 variants ORF contain 13 exons, spanning 2430 bp (variant 2–2427 bp) and encode a predicted protein of 810 amino acids (aa) (variant 2–809 aa) (Fig. 1).
Schematic representations of AtRAD21.1, AtRAD21.2, and AtRAD21.3 genes. Exons are shown as open boxes, and introns are shown as thick black lines. The 5′- and 3′-UTR of the three RAD21 genes is shown by a thick grey line. Black arrowheads represent primers used for expression analysis by RT-PCR (CR1, ACO5, ATRAD21-3-5, 5HOM2, 5CO-6, AT5). The T-DNA insertion sites in the AtRAD21 genes, atrad21.1 (salk_ 044851) and atrad21.3 (salk_076116) is shown by a large black triangle. The filled circles identify the putative separase target consensus sequence (S/D/xExxRx) (Hauf et al., 2001) in AtRAD21.1 that are predicted to be cleaved by the putative Arabidopsis separase. The numbers in the sequences depict the amino acid residue number in the protein sequence.
The cDNAs corresponding to AtRAD21.2 and AtRAD21.3 (Col-0) were also cloned and sequenced, along with their 5′- and 3′-UTR sequences. AtRAD21.2 has a 208 bp long 5′-UTR and a 185 bp 3-′UTR, and contains 12 exons and is 2079 bp long. AtRAD21.2 encodes a predicted protein of 693 aa (accession number AF400128) (Fig. 1).
AtRAD21.3 (Col-0) has a 227 bp long 5′-UTR and a 281 bp long 3′-UTR and is encoded by 15 exons and is 3093 bp long. AtRAD21.3 encodes a predicted protein of 1031 aa (accession number AF400129) (Fig. 1).
Sequence analysis of AtRAD21 proteins
The two variants of AtRAD21.1 differ by a single amino acid that does not significantly alter their predicted properties. PSORT analysis of AtRAD21.1 identified two potential nuclear localizing signals (NLS) (460RKRK463 and 490KRRNVPHTDCPERRTKRF507). AtRAD21.2 has two predicted NLS (355RKRK358 and 383RKRKK387) (PSORT). The largest of the three Arabidopsis RAD21 homologues, AtRAD21.3, has a single predicted NLS (546KRLRSAPRSTATKRKVL562; PSORT analysis). All three proteins have several phosphorylation sites each (PROSITE). These characteristics are in agreement with the putative cohesion function of these proteins. However, none of the AtRAD21 proteins identified here have predicted serine phosphorylation sites important for cohesin cleavage (Alexandru et al., 2001). Phosphorylation of serine residues in the ESP1 cleavage motif is necessary for the RAD21 cleavage that precedes loss of sister chromatid cohesion (Alexandru et al., 2001). Although it was possible to identify putative consensus ESP1 cleavage motifs in AtRAD21.1 (148SxExxRx154; 195DxExxRx201, and 218SxExxRx224) (Hauf et al., 2001) and in the meiotic cohesin homologue DIF1/SYN1 (Bai et al., 1999; Bhatt et al., 1999) (124DxExxRx130), similar putative ESP1 cleavage motifs were not identified in AtRAD21.2 and AtRAD21.3. If, however, the consensus sequence for ESP1 cleavage has diverged at the first amino acid S/T/D/I in the cleavage motif–/T/D/IxExxRx, there are several putative ESP1 cleavage motifs in AtRAD21.2 and AtRAD21.3. All AtRAD21 proteins have several PEST sequences (Rechsteiner and Rogers, 1996), suggesting that these proteins are likely to be unstable (Rao et al., 2001).
ClustalX1.8 alignment of the three AtRAD21 sequences with those of RAD21 proteins from other organisms show that the N and C termini are the most conserved (see supplementary Figs I and II at JXB online), as previously observed for RAD21 proteins identified in Arabidopsis and other organisms (McKay et al., 1996; Bai et al., 1999; Bhatt et al., 1999; Watanabe and Nurse, 1999; Warren et al., 2000). The conserved N and C terminal sequences of selected available plant, yeast, and animal REC8 and RAD21 were aligned with ClustalX1.8, and the data were used to generate a parsimony rooted tree with Phylip using AtSPO11-1 as the outgroup (see supplementary Fig. III at JXB online). Phylogenetic analysis of the three Arabidopsis RAD21 genes revealed that AtRAD21.2 (21AR2) and AtRAD21.3 (21AR3) both have closely related sequences in the rice genome, 21ORYZ3 and 21ORYZKAA, respectively, and occupy a position separate from DIF1/SYN1 (8AR) and the animal and yeast REC8 sequences. However, the relationship of AtRAD21.1 (21AR1) sequence to other RAD21 or REC8 protein sequences was not clear. Finally, the increase in number of RAD21 genes in relation to other model organisms is a common feature of both monocot and dicot genomes.
The three AtRAD21 genes are expressed in a wide variety of tissues
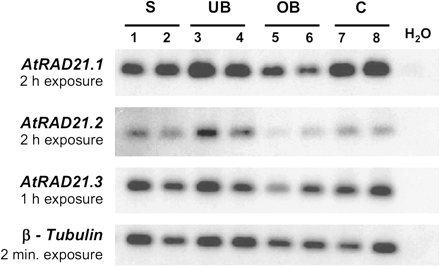
In order to determine and compare the expression profile of the three AtRAD21 genes, their expressions in different plant tissues were analysed. RT-PCR analysis of the three AtRAD21 genes shows that all three are expressed in tissues rich in cells undergoing mitosis, namely 7-d-old seedlings, unopened flower buds, open flower buds, and the rapidly proliferating inflorescence meristem of the ap1-1/cal-1 double mutant (Bowman, 1994) (Fig. 2). Although all three AtRAD21 genes show overlapping patterns of expression, their relative levels of expression are not similar. In plants grown under normal growth conditions AtRAD21.3 has the highest transcript level. It was possible to detect AtRAD21.3 transcripts in 5 μg of total RNA from tissues of untreated (non-irradiated) seedlings (Fig. 3), whereas AtRAD21.1 mRNA could only be detected in poly-A RNA samples of untreated inflorescences (not shown). A weak signal corresponding to the AtRAD21.2 transcript could be detected when 10 μg of total RNA from seedlings was used for northern blot analysis (Fig. 3).
RT-PCR expression analysis of the three AtRAD21 genes. AtRAD21 genes are expressed in vegetative and reproductive tissues containing dividing cells, but at different levels of expression. Autoradiograph representing the expression of all three genes, as analysed by RT-PCR using RNA extracted from 7-d-old seedlings (S), unopened flower buds (UB), open flowers (OB), and cauliflower-1/apetala1-1 inflorescence meristem tissue (C); a water control was also included in the analysis (H2O) and two independent products for each tissue are shown. Primers specific for AtRAD21.1, AtRAD21.2, AtRAD21.3, and the RT-PCR control – β-tubulin (Oppenheimer et al., 1988; Snustad et al., 1992; Knight et al., 1999) were used to amplify the corresponding cDNAs (Fig. 1) and 30 μl of each PCR reaction was loaded per well; these products were subsequently used for Southern hybridization with the corresponding radiolabelled probes to produce the autoradiographs shown. The analysis was performed with RT-PCR products from eight independent cDNA samples and a representative result is shown; each autoradiograph was exposed to the RT-PCR products for different periods.
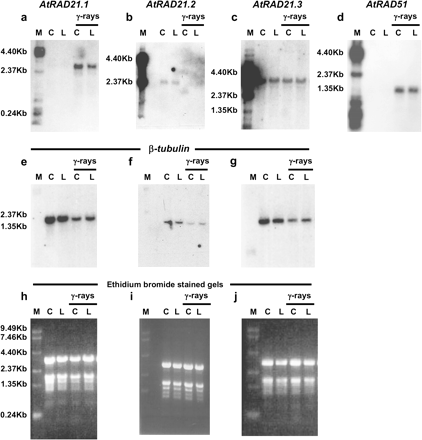
Transcript levels of AtRAD21.1 increase in plants exposed to ionizing radiation. The steady-state mRNA levels of AtRAD21.1 increase 1 h after ionizing radiation treatment, while AtRAD21.3 transcript levels remains mostly unchanged and AtRAD21.2 transcript levels are reduced. Autoradiographs show the steady-state RNA levels of AtRAD21.1 (a), AtRAD21.2 (b), AtRAD21.3 (c), AtRAD51 (d), and β-tubulin (e–g) in the aerial parts of 3-week-old seedlings exposed to 300 Gy of gamma rays (γ-rays) and in unexposed controls. Wild-type plants from Columbia-0 (C) and Landsberg erecta (L) ecotype were used for the analysis, and the ethidium bromide-stained gels showing equal loading are shown in (h)–(j). 5 μg of total RNA sample was loaded in each well, except for AtRAD21.2, for which 10 μg total RNA was used. RNA samples were extracted from aerial part of 3-week-old plants that had the following treatments: C, wild type Columbia ecotype (Col-0) non-irradiated; L, wild-type Landsberg ecotype (Lands.) non-irradiated; γ-rays C, Col-0 irradiated once with 300 Gy of gamma radiation; γ-rays L, Lands. irradiated once with 300 Gy of gamma radiation. The molecular weight marker (M) used was a 0.24–9.5 kb Gibco BRL RNA ladder. Autoradiographs for AtRAD21.1, AtRAD21.2, AtRAD21.3, and AtRAD51 were developed after exposure for 16 h, 14 d, 3 d and 16 h, respectively.
Ionizing radiation enhances AtRAD21.1 gene expression
Exposure of plants to DNA damage is known to increase transcript levels of genes implicated in DNA repair, and as rad21 mutants identified for several different organisms are also sensitive to γ-rays (Birkenbihl and Subramani, 1992; Heo et al., 1998; Losada et al., 1998; Sonoda et al., 2001), it was tested if γ-rays (and X-rays), expected to induce DNA DSB damage, could also alter the expression levels of any of the RAD21 genes in Arabidopsis.
There were specific differences in the expression of different AtRAD21 genes after exposure to ionizing radiation. Steady-state mRNA levels for AtRAD21.1 increase in the aerial tissues of wild-type plants 1 h after irradiation with ionizing radiation (γ-rays and X-rays) (Figs 3 and 4, respectively). The increase in AtRAD21.1 transcript after irradiation with γ-rays parallels the increased transcript levels of AtRAD51, a gene involved in the homologous recombination of broken double-stranded DNA (Klimyuk and Jones, 1997; Doutriaux et al., 1998). By contrast, steady-state mRNA levels for AtRAD21.2 were reduced in γ-ray irradiated plants, while the AtRAD21.3 expression pattern was not significantly altered after exposure to γ-rays (300 Gy). To determine if the response of AtRAD21.1 was specific to DNA DSB and not a broader response to DNA damage (ionizing radiation, X-rays and γ-rays, ultimately causes both double and single DNA strand breaks via oxidative stress), wild-type Arabidopsis plants were exposed to ultraviolet (UV-B) radiation. UV-B indirectly causes single-strand DNA breaks (rather than DSBs) through the formation of pyrimidine dimers and oxidative damage products which are substrates for excision repair pathways (Britt, 1999). It was found that AtRAD21.1 expression was not altered after plants were exposed to UV-B radiation (data not shown). This suggests that the increase in steady-state mRNA levels of AtRAD21.1 was a specific response to DSB DNA damage induced by ionizing radiation.
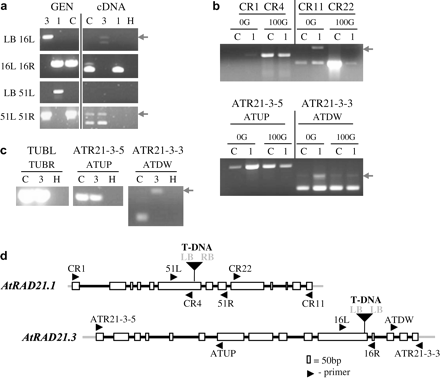
Characterization of the atrad21.1 and atrad21.3 mutant lines. T-DNA insertions in both atrad21.1 and atrad21.3 homozygous mutants truncate corresponding atrad21 transcripts. (a) PCR products indicating insertion of the T-DNA into the gene AtRAD21.3 (primer pair LB 16L) and AtRAD21.1 (primer pair LB 51L). The truncated atrad21.3 transcript detected in mutant atrad21.3 plants includes part of the T-DNA sequence. (b) top panel: Expression of the truncated atrad21.1 transcript is still increased after exposure of the atrad21.1 mutant to 100 Gy of ionizing radiation (X-rays), but this truncated transcript contains only sequences upstream of the T-DNA insertion site. The reduced expression detected downstream of the mutant atrad21.1 gene could be caused by a cryptic T-DNA promoter. Bottom panel: PCR control reaction with primers specific to the AtRAD21.3 gene showing that an equivalent amount of template from each sample was used in all four PCR reactions. (c) The atrad21.3 mutant expresses a truncated atrad21.3 transcript that includes sequences up to, and upstream of the T-DNA insertion site, but does not include sequences downstream of the T-DNA insertion site. β-tubulin primers used as control. In (a), (b), and (c) the arrow indicates the (top) contaminant genomic band. (d) Schematic representation of the T-DNA insertions and of the primers. LB and RB – T-DNA left and right border, respectively (in bold); primers for T-DNA left and right border (LB, RB); primers specific to AtRAD21.1 –CR1, CR4, 51L, 51R, CR22, CR11; primers specific to AtRAD21.3–ATRAD21-3-5, ATUP, 16L, 16R, ATDW, ATR21-3-3; primers for β-tubulin gene family (control) –TUBL, TUBR. Samples: genomic samples (GEN); cDNA of 5-d-old seedlings in (a) and (b); cDNA samples of inflorescences in (c). 1 (atrad21.1 homozygous mutant), 3 (atrad21.3 homozygous mutant); C (Columbia-0 plants). 0G and 100G (5-d-old seedlings samples non-irradiated and irradiated with 100 Gy of ionizing radiation (X-rays), respectively).
Mutants of AtRAD21.1, but not AtRAD21.3, are sensitive to ionizing radiation
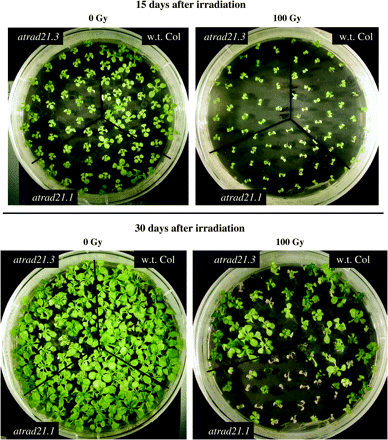
To assess the function provided by the AtRAD21 genes, lines with T-DNA insertions (Alonso et al, 2003) in AtRAD21.1 and AtRAD21.3 were characterized. The insertion mutant for AtRAD21.1 (salk_044851) has a T-DNA inserted in the 6th exon of the gene, likely to disrupt expression of AtRAD21.1. However, despite this insertion, atrad21.1 mutants have no obvious developmental phenotype. atrad21.1 mutant plants are fertile, produce viable seeds and undergo normal vegetative growth. Sequencing of the left and right borders of the T-DNA insertion site showed that the T-DNA insert is (Fig. 1) accompanied by an 8 bp deletion in the 6th exon of AtRAD21.1. Furthermore, AtRAD21.1 expression analysis in the homozygous mutant shows that the T-DNA insertion prevents expression of the full-length AtRAD21.1 transcript (Fig. 4). As AtRAD21.1 mRNA levels are increased after ionizing irradiation, it seems possible that AtRAD21.1 could have a role in repairing DNA damage and, consequently, loss of AtRAD21.1 may make mutant plants hypersensitive to DNA DSB damage. Therefore atrad21.1 mutants were exposed to DNA damage by growing seedlings on GM containing 1 μg ml−1 and 2 μg ml−1 of bleomycin (which causes single- and double-strand breaks in DNA), but did not observe any increase in sensitivity (data not shown). The radiation sensitivity of atrad21.1 and wild-type plants was also tested by exposing plants to X-rays at two different stages of development, either as seeds or as 5-d-old seedlings. Both seeds and seedlings were exposed to several different doses of X-rays (0, 50, 100, and 150 Gy) and their growth response was analysed after a fixed period. A hypersensitive response was consistently observed in atrad21.1 seeds exposed to sub-lethal doses of X-rays (Fig. 5); plants grown from irradiated atrad21.1 seeds also displayed a decrease in the number of true leaves and therefore showed a slight developmental delay when compared with seedlings germinated from wild-type irradiated seeds (see supplementary Fig. IV at JXB online). Surprisingly, when 5-d-old seedlings of atrad21.1 mutants were irradiated, rather than seeds, they did not show an increased sensitivity to radiation, compared with wild-type seedlings. This suggests that the function of AtRAD21.1 in DNA DSB repair is essential in seeds during imbibing/germination, but not in germinated seedlings.
Radiation hypersensitive phenotype of atrad21.1 mutants. Seeds of wild-type Columbia (Col-0) plants, and atrad21.1, and atrad21.3 mutants were irradiated (100 Gy) and germinated on GM medium. Growth of the irradiated seeds was compared to unirradiated samples (0 Gy) 15 and 30 d after treatment. No differences are seen 15 d after irradiation of w.t. Col (Col-0) and the two different atrad21 mutants, apart from the general delay in development of irradiated plants. However, a phenotype was evident when plants were allowed to grow longer, and analysed 30 d after irradiation, it was clear that atrad21.1 mutants were clearly more sensitive to irradiation than either atrad21.3 or Col-0, which supports a role for AtRAD21.1 in DNA DSB repair.
As AtRAD21.3 has the highest steady-state mRNA levels amongst all three AtRAD21 genes, it seemed likely that it might provide an essential mitotic function. Hence, a T-DNA insertion in AtRAD21.3 from the SALK T-DNA population (salk_076116) was also characterized. It was confirmed that the insertion of the T-DNA (with two left borders) in the 11th exon of AtRAD21.3 (Fig. 1) is associated with a 9 bp deletion. This insertion also disrupts AtRAD21.3 transcript integrity (Fig. 4). The loss of AtRAD21.3 function does not result in lethality or infertility, but mutant plants develop slightly slower than wild type and, consequently, start to flower (bolting) a bit later than the wild type (data not shown). As AtRAD21.3 transcript persists after irradiation with γ-rays, atrad21.3 plants were also tested for sensitivity to DNA damage by exposing them (seeds and seedlings) to X-rays, or to 1 μg ml−1 and 2 μg ml−1 of bleomycin, respectively, and no difference was found in the bleomycin and radiation sensitivity of wild-type and mutant plants (Fig. 5) (data not shown for the bleomycin assay).
The DIF1 promoter drives expression in reproductive and vegetative tissues
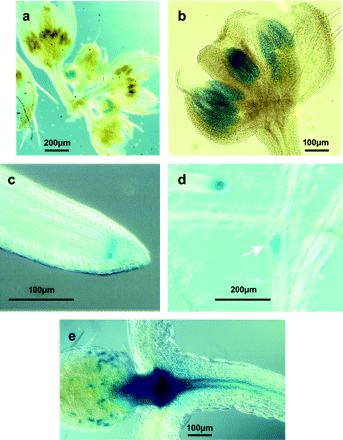
DIF1/SYN1, the Arabidopsis RAD21 homologue essential only for meiosis, has been shown by RT-PCR to be expressed not only in flowers but also in vegetative tissue, namely seedlings (Bai et al., 1999; Bhatt et al., 1999). Therefore, in order to determine if DIF1 expression in seedlings correlates with actively dividing mitotic cells, and hence if its expression overlaps with that expected of the other AtRAD21 genes, the expression pattern of the 520 bp upstream sequence of DIF1 fused to the uidA reporter gene, encoding β-glucuronidase (GUS) was investigated. This fragment contains the 5′-end of the DIF1/SYN1 transcript that overlaps with a flanking divergently transcribed ORF of a glycoamidase-like gene (Bai et al., 1999), and hence it is likely that this 520 bp promoter fragment retains elements important for DIF1 expression. 26 independent transformants were generated and the tissue- and stage-specific expression of this reporter was examined in a subset of these, of which a representative sample is shown in Fig. 6. Of these 26 transformants, 11 stained in the young anther and root tips, 19 in the emerging lateral roots, and 21 in the shoot apical meristem region. Nine displayed a puntacted-staining pattern in the basal part of the young leaves. As expected from its function during meiosis in anthers and ovules, reporter gene expression controlled by the DIF1 promoter was seen in the anthers of flower buds at stages 7–10 (staging according to Smyth et al., 1990) (Fig. 6a, b), however, it was not possible to detect reporter expression in ovules. In addition to the expression in reproductive organs, GUS expression was also seen in the root tips (Fig. 6c), in emerging lateral roots (Fig. 6d) and at the shoot apex (Fig. 6e). The reporter gene was also expressed in a punctuate pattern in developing leaves (Fig. 6e). This suggests that DIF1 expression may be associated with dividing cells in the shoot apexes and vegetative tissue, and may possibly overlap with the expression of the AtRAD21 homologues.
DIF1 promoter–uidA expression pattern is not restricted to flowers. The DIF1 promoter–-uidA fusion was detected using a chromogenic substrate. The reporter gene was expressed in buds at inflorescences stages 7–10 (a), specifically in the anthers (b); expression was also seen in the root-tip (c) and in emerging lateral roots (d) of 7-d-old transgenic seedlings. The β-glucuronidase protein encoded by the uidA reporter gene was also detected in young leaves (e) and around the shoot apex (e) of 7–10-d-old seedlings. Expression of DIF1 could hence overlap the expression of the AtRAD21 genes in vegetative meristematic tissues.
AtRAD21.1 and AtRAD21.3 share redundant functions
Since two of the AtRAD21 genes appear to be at least partially redundant for normal growth, attempts were made to isolate a double mutant to determine if this would result in a vegetative or reproductive phenotype, or affect the viability of a atrad21.1 atrad21.3 double mutant. However, the atrad21.1 atrad21.3 double mutants are normal in all aspects of their development and are fully fertile with normal seed set (data not shown). Thus, to unmask the mitotic function of RAD21 genes in Arabidopsis may require the generation of plants mutant for three or more RAD21/REC8 genes.
Discussion
Structural features of the three Arabidopsis AtRAD21 proteins
Most organisms have been reported to have two RAD21 homologous genes, a meiotic and a mitotic one (Birkenbihl and Subramani, 1992; Molnar et al., 1995; McKay et al., 1996; Guacci et al., 1997; Michaelis et al., 1997; Heo et al., 1998; Losada et al., 1998; Klein et al., 1999; Watanabe and Nurse, 1999; Warren et al., 2000). In this study, it is shown that A. thaliana, like C. elegans (Pasierbek et al., 2001) and rice, has three RAD21 homologues in addition to the meiotic REC8 homologue (Bai et al., 1999; Bhatt et al., 1999; Pasierbek et al., 2001).
All three AtRAD21 proteins have NLS, as would be expected of proteins likely to be involved in cohesion of sister chromatids (Birkenbihl and Subramani, 1995; Uhlmann and Nasmyth, 1998; Losada and Hirano, 2001). All three AtRAD21 proteins are also expected to be unstable as they have potential PEST sequences and are predicted to have several phosphorylation sites that may be important for their function. In yeast, RAD21 is hyperphosphorylated and is subsequently rapidly degraded to release sister chromatid cohesion (Birkenbihl and Subramani, 1995; Uhlmann et al., 1999, 2000; Tomonaga et al., 2000). In addition to these sites, the presence of putative separase cleavage sites in AtRAD21.1, originally identified for SCC1 (a RAD21 homologue) (Uhlmann et al., 2000; Hauf et al., 2001), suggests that AtRAD21.1 may also be subjected to regulation via an ESP1 cleavage pathway. Although consensus ESP1 cleavage sites were not identified in the AtRAD21.2 and AtRAD21.3 proteins, it is possible that these plant RAD21 proteins may have a slightly different ESP1 cleavage site from those based on the animal and yeast consensus sequences (Fig. 1). As Arabidopsis has a putative ESP1 homologue (Jones and Sgouros, 2001; At4g22970), it is speculated that the protease-mediated putative cleavage of Arabidopsis RAD21 proteins is also likely to be a feature involved in the regulation of their function. Thus, the mechanism of chromosome cohesion release via ESP1 cleavage of cohesion which is conserved in other organisms could also be conserved in Arabidopsis.
The AtRAD21.1 protein is involved in DSB DNA repair and may share redundant functions with other family members during cell division
In Arabidopsis, RT-PCR analysis of the expression for three AtRAD21 genes shows that all three (AtRAD21.1 to AtRAD21.3) are expressed in tissues with dividing cells (Fig. 2), as would be expected for genes predicted to be involved in chromosome cohesion. As DIF1/SYN1 (REC8 homologue) is expressed in vegetative and reproductive cells (Bai et al., 1999; Bhatt et al., 1999), the overlapping expression pattern of the REC8/RAD21 genes in Arabidopsis could thus provide redundancy in one or more functions at different stages of development, namely in meristematic tissue. This appears be the case as the atrad21.1 atrad21.3 double mutant develops normally and is fully fertile, as would be expected for genes that are at least partially redundant.
For one of the Arabidopsis RAD21 genes, AtRAD21.1, an increase in expression is specifically induced by both ionizing radiation (this study) and radiation mimic agents (West et al., 2004), suggesting a role in DNA DSB repair. Furthermore, expression data from microarray (Affymetrix25K – www.arabidopsis.org/servlets microarray database) Genevestigator (https://www.genevestigator.ethz.ch/; Zimmermann et al., 2004), and AtGenExpress (http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm) emphasize that the increase in AtRAD21.1 gene expression after exposure to ionizing radiation is probably induced directly by DNA DSB damage rather than by oxidative damage (caused by ionizing radiation) or other types DNA damage; exposure to oxidative stress per se, UV-A, UV-B, and UV-A+B do not lead to an increase of AtRAD21.1 gene expression.
Although this study's analysis does not demonstrate that the increase in AtRAD21.1 transcript results in a corresponding increase in the pool of AtRAD21.1 protein at sites of DNA damage, similar to that described in yeast (Ünal et al., 2004; Ström et al., 2004), the hypersensitivity of atrad21.1 mutants to ionizing radiation inducing DNA DSB damage and its expression enhancement as an emergency response to ionizing (and ionizing mimicking agents) suggest a role for AtRAD21.1 in DNA repair, possibly by facilitating repair between homologous sister chromatids. As the radiation-hypersensitive phenotype was only observed in atrad21.1 plants that developed from irradiated seeds, but not in mutant plants that were irradiated after germination, it is speculated that expression of AtRAD21.1 in the irradiated seeds is critical for a normal response to DNA damage. It is known that seed desiccation results in genome damage, which requires DNA repair during the seed imbibing/germination stage (Britt, 1996; Dandoy et al., 1987). Finally, during embryogenesis there is a 5–10-fold increase in gene expression of AtRAD21.1 while the amounts of the other AtRAD21 transcripts remain unaltered in the later stages of embryo development (microarray data from AtGenExpress: http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm); these data, taken in conjunction with the radiation-hypersensitive phenotype of atrad21.21 mutants and the increase in AtRAD21.1 transcripts by radiation damage, support such a role for AtRAD21.1 in the repair of DNA damage specifically during seed imbibing/germination. Irradiated atrad21.1 seedlings fail to show radiation hypersensitivity, possibly because one of the other RAD21 homologues could provide redundancy at this stage.
The increase in steady-state mRNA levels of AtRAD21.1 after γ-irradiation of Arabidopsis is not unique, as it has also been observed for several other genes like MIM, AtLigIV, AtGR1, AtRAD51, AtBRCA1, and AtKu80 after exposure to DNA DSB inducing agents (Klimyuk and Jones, 1997; Doutriaux et al., 1998; Mengiste et al., 1999; Deveaux et al., 2000; Hanin et al., 2000; West et al., 2000; Friesner and Britt, 2003; Gallego et al., 2003; Lafarge and Montané, 2003). These genes are involved in either homologueous recombination (HR) or non-homologous end joining (NHEJ) DNA repair of DNA DSB. Of these, mutants for MIM, AtLigIV, AtKu80, like atrad21.1 mutants, display a hypersensitive phenotype after exposure to DSB-inducing agents.
Apart from the slower bolting phenotype of the atrad21.3 mutants (salk_076116) no other vegetative, or reproductive defects could be identified, probably due to the function provided by the two other AtRAD21 genes, or that of the REC8 homologue DIF1/SYN1 in vegetative tissues. Since AtRAD21.3 is expressed at high levels and is not altered by radiation, it may be involved in basic structural roles, perhaps associated with the nuclear matrix and/or sister chromatid cohesion (Sadano et al., 2000). If this is the case, its possible role in chromosome structural maintenance could explain the general delay in development, especially the delayed bolting phenotype of the atrad21.3 mutant.
Our analysis of atrad21.1 and atrad21.3 single and double mutants suggests that the functions provided by these two AtRAD21 genes are redundant with that of other RAD21/REC8 family members for vegetative development and fecundity of Arabidopsis. It is possible that, apart from these two AtRAD21 genes, either AtRAD21.2, or the REC8 homologue DIF1/SYN1, both of which are expressed in overlapping domains with AtRAD21.1 and AtRAD21.3 could provide such redundant function (Bai et al., 1999; Bhatt et al., 1999; Dong et al., 2001). However, AtRAD21.1 does have a specific function in DNA repair, evident only after mutant seeds are exposed to ionizing radiation, which is not compensated for by the expression of AtRAD21.3. This is in accordance with the fact that it was not possible to identify a vegetative phenotype in atrad21.1 atrad21.3 double mutants. The analysis of Arabidopsis RAD21 genes in mitosis may require the generation of plants mutant for all three RAD21 genes and/or a quadruple mutant deficient in RAD21 and REC8 functions. This should reveal if indeed AtRAD21 proteins have any role in sister chromatid cohesion during mitosis or meiosis.
Accession numbers
The sequences of the AtRAD21 genes (Col-0) have the following NCBI accession numbers: AF400126 (AtRAD21.1 variant 1), AF400127 (AtRAD21.1 variant 2), AF400128 (AtRAD21.2), and AF400129 (AtRAD21.3).
We are most thankful to Dr I Moore (for advice), Professor Dr P Cook (γ-rays source; University of Oxford, UK), Drs S Ferrari and M El-Shemerly (X-ray source; University of Zürich, Switzerland), and to Dr P Talhinhas (ISA, Lisboa, Portugal) for his help on phylogeny tree construction. We are also grateful to the program PRAXIS XXI (Portugal) for providing the funding (scholarship PRAXIS XXI/BD/9484/96) to JA da C-N. Funding in HGD's laboratory is provided by the Biotechnology and Biological Sciences Research Council, UK, and in UG's laboratory is provided by the Swiss National Science Foundation and the University of Zürich.
References
Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K.
Alonso JM, Stepanova AN, Leisse TJ, et al.
Bai X, Peirson BN, Dong F, Xue C, Makaroff C.
Batoko H, Zheng HQ, Hawes C, Moore I.
Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones G, Dickinson H, Dean C.
Birkenbihl RP, Subramani S.
Birkenbihl RP, Subramani S.
Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE, Vidal M.
Bowman J.
Britt A.
Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, Meyer BJ.
Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Tóth A, Shevchenko A, Nasmyth K.
Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K.
Clough SJ, Bent AF.
Dandoy E, Schyns R, Deltour L, Verly WG.
Deveaux Y, Alonso B, Pierrugues O, Godon C, Kazmaier M.
Dong F, Cai X, Makaroff CA.
Doutriaux MP, Couteau F, Bergounioux C, White C.
Edwards K, Johnstone C, Thompson C.
Felsenstein J.
Friesner J, Britt A.
Gallego ME, Bleuyard JY, Daoudal-Cotterell S, Jallut N, White CI.
Guacci V, Koshland D, Strunnikov A.
Hanin M, Mengiste T, Bogucki A, Paszkowski J.
Hauf S, Waizenegger IC, Peters JM.
Heo S, Tatebayashi K, Kato J, Ikeda H.
Jefferson RA, Kavanagh TA, Bevan MW.
Jones JD, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K.
Jones S, Sgouros J.
Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K.
Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K.
Klimyuk VI, Jones JDG.
Knight H, Veale E, Warren GJ, Knight MR.
Lafarge S, Montané MH.
Logemann J, Schell J, Willimitzer L.
Losada A, Hirano T.
Losada A, Hirano M, Hirano T.
Losada A, Yokoshi T, Kobayashi R, Hirano T.
Mayer ML, Gygi SP, Aebersold R, Hieter P.
McElver J, Tzafrir I, Aux G, et al.
McKay MJ, Troelstra C, van der Spek P, Kanaar R, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers JHJ.
Mengiste T, Revenkova E, Bechtold N, Paszkowski J.
Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin CH.
Michaelis C, Ciosk R, Nasmyth K.
Mito Y, Sugimoto A, Yamamoto M.
Molnar M, Bähler J, Sipiczki M, Kohli J.
Oppenheimer DG, Haas N, Silflow CD, Snustad DP.
Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J.
Person WR, Wood T, Zhang Z, Miller W.
Rao H, Uhlmann F, Nasmyth K, Varshavsky A.
Rechsteiner M, Rogers SW.
Rodrigues-Pousada RA, De Rycke R, Dedonder A, Caeneghem WV, Engler G, Van Montagu M, Van Der Straeten D.
Sambrook J, Fritsch EF, Maniatis T.
Sadano H, Sugimoto H, Sakai F, Nomura N, Osumi T.
Sjögren C, Nasmyth K.
Skibbens RV, Corson LB, Koshland D, Hieter P.
Smyth DR, Bowman JL, Meyerowitz EM.
Snustad DP, Haas NA, Kopczak SD, Silflow CD.
Sonoda E, Matsusaka T, Morrison C, et al.
Ström L, Lindroos HB, Shirahige K, Sjögren C.
Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM.
The Arabidopsis Genome Initiative.
Tomonaga T, Nagao K, Kawasaki Y, et al.
Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K.
Uhlmann F, Lottspeich F, Nasmyth K.
Uhlmann F, Nasmyth K.
Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K.
Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D.
Waizenegger IC, Hauf S, Meinke A, Peters JM.
Warren WD, Lin E, Nheu TV, Hime GR, McKay MJ.
Watanabe Y, Nurse P.
West CE, Waterworth WM, Jiang Q, Bray CM.
West CE, Waterworth WM, Story GW, Sunderland PA, Jiang Q, Bray CM.
West CE, Waterworth WM, Sunderland PA, Bray CM.




Comments