-
PDF
- Split View
-
Views
-
Cite
Cite
Tyrone C. Spady, Ole Seehausen, Ellis R. Loew, Rebecca C. Jordan, Thomas D. Kocher, Karen L. Carleton, Adaptive Molecular Evolution in the Opsin Genes of Rapidly Speciating Cichlid Species, Molecular Biology and Evolution, Volume 22, Issue 6, June 2005, Pages 1412–1422, https://doi.org/10.1093/molbev/msi137
Close - Share Icon Share
Abstract
Cichlid fish inhabit a diverse range of environments that vary in the spectral content of light available for vision. These differences should result in adaptive selective pressure on the genes involved in visual sensitivity, the opsin genes. This study examines the evidence for differential adaptive molecular evolution in East African cichlid opsin genes due to gross differences in environmental light conditions. First, we characterize the selective regime experienced by cichlid opsin genes using a likelihood ratio test format, comparing likelihood models with different constraints on the relative rates of amino acid substitution, across sites. Second, we compare turbid and clear lineages to determine if there is evidence of differences in relative rates of substitution. Third, we present evidence of functional diversification and its relationship to the photic environment among cichlid opsin genes. We report statistical evidence of positive selection in all cichlid opsin genes, except short wavelength–sensitive 1 and short wavelength–sensitive 2b. In all genes predicted to be under positive selection, except short wavelength–sensitive 2a, we find differences in selective pressure between turbid and clear lineages. Potential spectral tuning sites are variable among all cichlid opsin genes; however, patterns of substitution consistent with photic environment–driven evolution of opsin genes are observed only for short wavelength–sensitive 1 opsin genes. This study identifies a number of promising candidate-tuning sites for future study by site-directed mutagenesis. This work also begins to demonstrate the molecular evolutionary dynamics of cichlid visual sensitivity and its relationship to the photic environment.
Introduction
Visual ecologists have long observed a correlation between the photic environment and visual sensitivity (Bowmaker 1995). Some of the most dramatic examples have been found in fish rod photoreceptors. Deep-sea fish rod spectral sensitivities are shortwave shifted, relative to shallow-dwelling species, to match the ambient spectra of the deep-sea environment (Partridge, Archer, and Lythgoe 1988, Crescitelli 1991). Adaptation to depth has also been observed in the rods of freshwater teleosts. Among the cottoids of Lake Baikal, the world's deepest lake, rhodopsin's (Rh1) absorption maxima decreases as depth increases (Hunt et. al. 1996). Muntz (1976) compared a shallow and deeper living pair of closely related cichlid species of the genus Lethrinops. Again, the deepwater species had shortwave-shifted rod sensitivity.
There are also several examples of differences in cone spectral sensitivity associated with disparities in photic environment. Both deep-dwelling Lake Baikal cottoids and coelacanths show a marked shortwave shift in cone spectral sensitivities (Bowmaker et al. 1994; Yokoyama et al. 1999). Lutjanid fishes, of the Great Barrier Reef, demonstrate the interaction between water clarity and cone spectral sensitivity, with fish in clearer habitats having shortwave-shifted visual sensitivities (Lythgoe et al. 1994).
Visual pigments determine spectral sensitivity and are spectrally distinct photosensory molecules in the outer segments of retinal photoreceptor cells. Visual pigments are composed of a vitamin A–derived chromophore bound to an opsin protein. Photoisomerization of the chromophore initiates the transductional cascade culminating in a neural response. Interactions between the chromophore and the amino acid residues of the opsin protein determine the absorbance properties of a visual pigment (Sakmar, Franke, and Khorana 1989; Zhukovsky and Oprian 1989; Nathans 1990a, 1990b; Sakmar et al. 2002; Yokoyama 2002).
Spectral sensitivity of cichlid fishes can be tuned using four nonexclusive mechanisms. First, the lenses of some cichlids contain inert short wavelength–absorbing carotenoid pigments (Thorpe, Douglas, and Truscott 1993). The presence of ocular pigments seems to be independent of photic environment, with nonpigmented species occurring in fish of both turbid and clear habitats (Thorpe, Douglas, and Truscott 1993).
Second, Carleton and Kocher (2001) have shown that cichlids use differential cone opsin expression to modulate visual sensitivity. Cichlids have six opsin genes (five cone opsins and one rod opsin): long wavelength–sensitive (LWS), rhodopsin-like (Rh2), short wavelength–sensitive 2b (SWS2b), short wavelength–sensitive 2a (SWS2a), short wavelength–sensitive 1 (SWS1), and rod opsin, rhodopsin (Rh1). Individual species express varying subsets of the five cone opsin genes. For example, the ambush predator Dimidiochromis compressiceps expresses LWS, Rh2, and SWS2a genes. In contrast, the planktivorous Metriaclima zebra expresses Rh2, SWS2b, and SWS1, a radically different subset of opsin genes.
Third, chromophore usage can vary among cichlids (vitamin A1 or A2 derived). Visual pigments based on a vitamin A2–derived chromophore have long wavelength–shifted absorbance maxima, relative to those based on a vitamin A1–derived chromophore (Partridge and Cummings 1999). Fish that inhabit turbid environments more commonly use A2 or A1-A2 mixtures (Bowmaker 1995), although chromophore thermal stability may also shape usage (Partridge and Cummings 1999). Cichlids from the turbid waters of Lake Victoria utilize A1-A2 mixtures (van der Meer and Bowmaker 1995); however, clear-water Lake Malawi cichlids use only A1 chromophores (Carleton, Harosi, and Kocher 2000; R. C. Jordan, K. A. Kellog, F. Juanes, J. R. Stanffer, and E. R. Loew, unpublished data).
Finally, amino acid substitutions in the opsin protein can alter visual sensitivity (summarized in Yokoyama 2002 and Takahashi and Ebrey 2003). The effects of individual substitutions are highly variable, ranging from 0 to 75 nm. Further, the effects of individual substitutions depend upon the amino acid background of the opsin protein.
There is mounting evidence for molecular adaptation to photic environment, via amino acid substitutions in opsin proteins, in East African cichlids. Recently, Sugawara, Terai, and Okada (2002) found evidence of functional divergence in Tanganyikan cichlid Rh1 opsin genes. They found an A292S substitution in several Tanganyikan lineages (bovine rhodopsin numbering will be used exclusively in this paper). In mammalian LWS visual pigments, an A292S substitution causes a −18-nm spectral shift and can be expected to have a similar effect in cichlid Rh1 visual pigments. Interestingly, all three species with the A292S substitution are deepwater species, further supporting the relationship of spectral sensitivity to depth. Further, Terai et al. (2002) report high variation of the LWS gene in Lake Victoria cichlids and highlight several potential functionally important substitutions (reviewed in Carleton and Kocher 2003). Terai et al. (2002) contend that similarities between ancestral and modern photic environment maintained ancestral variation in the Lake Victoria LWS gene.
The present study looks for evidence of adaptive molecular evolution among closely related cichlid species that inhabit dramatically different photic environments. We then test for differences in relative rates of evolution between turbid and clear-water lineages. Finally, we identify amino acid substitutions that are likely to be involved in the adaptive-functional differentiation of cichlid opsins. Cichlids endemic to clear lakes Tanganyika and Malawi are contrasted against cichlids endemic to more turbid environments in Lake Victoria and the Nile River. Due to geologic and climatic conditions, Lake Tanganyika and Lake Malawi are among the clearest freshwater systems in the world (Muntz 1976) and provide a stark contrast to the generally more turbid environments of Lake Victoria (Seehausen, van Alphen, and Witte 1997) and the Nile River. Because turbidity directly limits the transmission of the shortest wavelengths of the visible spectrum, the spectrum of ambient light is shifted toward the long-wavelength region. Thus, the spectral breadth of light available for vision is restricted in turbid habitats.
We use Codon-based Maximum Likelihood methods (CodeML; Yang et al. 2000), as implemented in Phylogenetic Analysis by Maximum Likelihood (PAML; Yang 1997), to look for evidence of adaptive molecular evolution. Comparisons of nonsynonymous (dN) and synonymous substitution rates (dS), dN/dS = ω, are used to infer the selective regime experienced by a gene (Graur and Li 2000). When ω = 1, a neutral mode of evolution is indicated. ω < 1 indicates purifying selection. ω > 1 indicates positive selection. PAML methods account for the different functional and structural constraints experienced by individual sites-domains of a protein by allowing for heterogeneous ω across sites (i.e., Yang and Swanson 2002). PAML has been used to detect positive selection among fertilization proteins (Civetta 2003; Swanson, Nielson, and Yang 2003), lysozymes (Yang 1998; Yang and Nielsen 2002), tumor suppressors (Yang and Nielsen 2002), dopamine receptors (Ding et al. 2002), and among insect opsins (Briscoe 2001).
Methods
Polymerase Chain Reaction and Sequence Analysis
We sequenced SWS1, SWS2a, SWS2b, Rh2, LWS, and Rh1 cichlid opsin genes. Opsin-coding sequences were obtained for 17 East African cichlid species (table 1). Cone opsin gene sequences from five species and rod opsin gene sequences from two of those species were obtained from GenBank (Carleton, Harosi, and Kocher 2000; Carleton and Kocher 2001; K. L. Carleton, J. W. L. Parry, J. K. Bowmaker, D. M. Hunt, and O. Seehausen, in preparation). All other rod opsin sequences and the cone opsin sequences from the remaining 12 species are new additions to the sequence database. The species studied represent lineages from the Nile River and Lake Victoria, both turbid, and Lakes Malawi and Lake Tanganyika, both clear. Retinal tissue was used to extract opsin messenger RNA, whenever possible. Retinas were homogenized and RNA extracted with Trizol (Invitrogen, Carlsbad, Calif). Retinal RNA preparations were then reverse transcribed with a poly T primer and Superscript II Reverse Transcriptase (Invitrogen). Genomic DNA was extracted as well and used to determine opsin-coding sequences when necessary. Opsin-coding sequences were polymerase chain reaction (PCR) amplified using sequence-specific primers as previously described by Carleton and Kocher (2001) for each of the six opsin classes found in cichlids SWS1, SWS2a, SWS2b, Rh1, Rh2, and LWS. Dynazyme Ext. (MJ Research, Waltham, Mass.), a polymerase mixture containing a high-fidelity polymerase with 3′–5′ proofreading activity, was used to amplify all sequencing templates. Opsin PCR products were sequenced using previously described primers and a DYEnamic™ ET terminator cycle sequencing kit (Amersham Pharmacia Biotech, Piscataway, N.J.) (Carleton and Kocher 2001). Sequences were aligned using Sequencher 4.1.2 (Gene Codes Corporation, Ann Arbor, Mich.). Whenever possible the complete opsin-coding sequence was used. In cases where complete sequences were not obtained, at least 97% of the continuous coding sequence from each opsin gene (6) was used. Regions of missing sequence, made up of small stretches in the 3′ and/or 5′ part of the coding sequence, were not used in the analysis. The regions used in the analysis always included all transmembrane (TM) and inter-TM regions, which control the spectral absorbance of visual pigments.
Study Species
Study Species
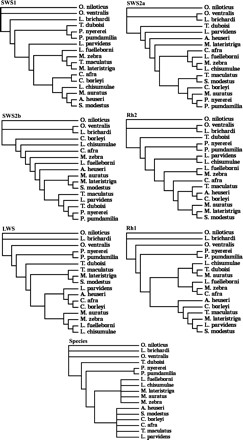
Gene trees were generated from opsin-coding sequences. Using PAUP (Swofford 2002), aligned sequences were used to calculate bootstrap trees (100 replicates, 50% majority rule). Bootstrap topologies were then used as a constraint in maximum likelihood estimation of gamma parameters. Maximum likelihood estimates of gamma parameters and Tamura-Nei distances were then used to generate unrooted neighbor-joining (Saitou and Nei 1987) tree topologies (fig. 1). Additionally, a putative species tree was constructed based on published cichlid phylogenies (Kocher et al. 1995, Streelman et al. 1998; Albertson et al. 1999).
Unrooted neighbor-joining tree topologies were generated based on Tamura-Nei distances and maximum likelihood estimates of gamma-shape parameters. Additionally, both the putative species tree and star tree were used in the analysis.
Maximum Likelihood Analysis
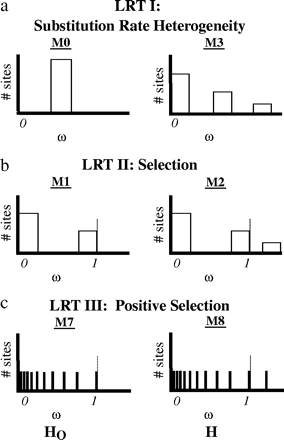
A total of seven tree topologies were used by PAML to examine relative rates of substitution. Each opsin gene was tested with all gene trees and a putative species tree. Nested models were compared using a likelihood ratio test (LRT) as described in Yang et al. (2000) and Yang and Nielsen (2002). The LRT statistic was calculated as twice the difference in maximum likelihood values (2Δℓ) between nested models. The significance of the LRT statistic was determined using a χ2 distribution. The standard degrees of freedom were used for each analysis (i.e., Yang and Nielsen 2002). Several site-specific likelihood models were used to fit the data. These models allow for ω heterogeneity among sites, except in the case of the one rate model. In all models, synonymous rates of substitution are assumed to be invariant across sites. Three LRTs were carried out using site-specific models (fig. 2). (1) The comparison of M0 (one rate) and M3 (discrete) was used to test for rate heterogeneity among amino acid sites (fig. 2a). M0 (one rate) averages the rates of substitution across all sites. M3 (discrete) assumes rate variation by allowing for a discrete number of rate categories (three used here). (2) The comparison of M1 (neutral) and M2 (selection) was used to test for selection (fig. 2b). M1 (neutral) allows for two rate classes, one with ω = 0 and the other with ω = 1. M2 is a variant of M1 and adds an additional unconstrained rate class where ω can be greater than 1. The stringent nature of M2 allows for either a class of sites under weak purifying selection or positive selection. M2 likelihood optimization can be affected by local optima (Yang et al. 2000; Anisimova, Bielawski, and Yang 2001; Wong et al. 2004). To alleviate this issue, starting ω values both above and below one were used. (3) The comparison of M7 (beta) and M8 (beta&ω) was used to test for positive selection (fig. 2c). These models allow for more continuous variation in substitution rates across sites. In both M7 and M8, there are 10 rate classes, of a fixed proportion of site, constrained to ω ≤ 1, where the shape of the distribution is defined by an additional parameter, beta. In M8 (beta &ω), an additional rate class is unconstrained. Again, to compensate for local optima effects, starting ω values both above and below one were used.
PAML calculates a maximum likelihood value (ℓ) for each model (for each gene). Nested models are compared using a LRT (LRT = 2Δℓ). The significance of the LRT is determined using a χ2 distribution, where the degrees of freedom are equal to 4, 2, and 2, respectively, for each test (see below). (a) The comparison of M0 (one rate) and M3 (discrete) was used to test for rate heterogeneity among amino acid sites. (b) The comparison of M1 (neutral) and M2 (selection) was used to test for positive selection. (c) The comparison of M7 (beta) and M8 (beta&ω) was used to test for positive selection.
Yang and Nielsen's Model B (2002) was used to test for differences between clear and turbid water lineages. Model B is a derivative of M3 (discrete), which simultaneously allows for site and branch heterogeneity in relative substitution rates. First, both turbid lineages were simultaneously compared to the clear-water lineages. Each turbid lineage was then tested individually with the other turbid lineage removed from the analysis. M3 (discrete) served as the null hypothesis for all Model B comparisons.
Site Predictions
Sites that are likely to be involved in the functional differentiation of cichlid opsin genes are determined using two methods. First, PAML uses an empirical Bayes approach to identify amino acid sites that are likely to have been under positive selection. Second, previously published structural and functional studies are used to predict functionally important substitutions. All variable sites were mapped onto the rhodopsin crystal structure (1L9H) (Palczewski et al. 2000; Teller et al. 2001). Functionally relevant sites are typically in the chromophore-binding pocket and often involve a change in amino acid polarity. While variation at a binding pocket site does not prove changes in spectral sensitivity, nearly all known spectral tuning sites are in the retinal-binding pocket (Yokoyama 2002; Takahashi and Ebrey 2003).
Results
Opsin Sequences
Consistent with the close phylogenetic relationships of the species sampled, most nucleotide sites were invariant in pair wise comparisons. In total, however, approximately 5%–10% of sites were variable, out of an average 1,035 bp of sequence for each gene. The least variation was seen amongst SWS2a and Rh1, with 4.7% and 5.1% variable nucleotide sites, respectively. SWS2b, Rh2, and LWS had intermediate proportions of variable nucleotide sites, at 6.8%, 7.5%, and 7.8%, respectively. SWS1 had the highest variation, with 10.5% of nucleotide sites being variable. Interestingly, Neolamprologus brichardi and Tropheus duboisi have accumulated frameshift mutations in SWS1 (90) and SWS2a (253), and SWS2b (97), respectively. However, when translated without frameshift mutations, we do not observe an excess of substitutions for any of the pseudogenes, which suggests that the nonfunctionalization has been fairly recent. Further, both loci were homozygous in the individuals sequenced. This suggests that these differences may be fixed or at least at high frequency.
Phylogenetic Reconstructions
The small number of substitutions among the 17 species used in this analysis minimized the need for multiple hit corrections of sequence divergence. Several substitution models were implemented and each gave very similar divergence estimates (data not shown). The Tamura-Nei model was chosen for subsequent analyses.
In their simulation study, Anisimova, Bielawski, and Yang (2001) detected positive selection using the LRT for tree lengths as small as 0.11 (nucleotide substitutions per codon along the tree). They not only documented the conservative nature of the LRT at low tree lengths but also showed that this can be remedied by increasing the number of sequences analyzed. Tree lengths varied among the genes analyzed in this study. The SWS1 tree had the largest tree length of 0.39. The SWS2a tree had the smallest tree length of 0.16. SWS2b, Rh1, Rh2, and LWS had intermediate tree lengths of 0.24, 0.23, 0.28, and 0.29, respectively.
Overall tree topologies were largely conserved among unrooted gene trees and were consistent with published East African cichlid phylogenies based on mitochondrial, microsatellite, and nuclear markers (Kocher et al. 1995, Streelman et al. 1998; Albertson et al. 1999) (fig. 1). Among the Lake Malawi species in particular, phylogenetic relationships were highly variable. Rock and sand dwellers were usually intermingled except in the Rh1 tree. The lack of perfect agreement among cichlid opsin gene trees is consistent with observations by other investigators that ancestral polymorphisms have not completely sorted among these species (Moran and Kornfield 1993). Because of this, the phylogenetic relationships reconstructed from single-gene loci may not reflect relationships at other loci or at the organismal level (Pamilo and Nei 1988; Streelman et al. 1998; Albertson et al. 1999; Kocher 2003). It is for this reason that several gene trees are used in this study, and we do not rely solely on a consensus tree or putative species tree, as other researchers have done (Sugawara, Terai, and Okada 2002).
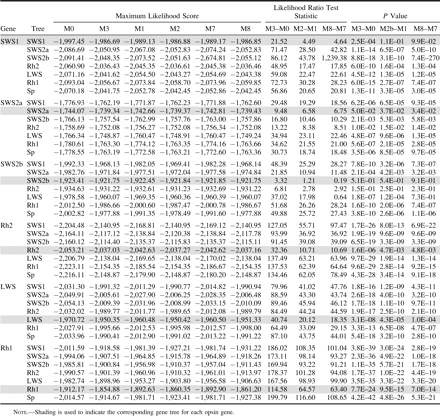
Yang et al. (2000) and Ford (2001) have asserted that mild uncertainty in tree topology only has a limited effect on the LRT. However, to test for the effects of tree topology, we tested each gene with the corresponding gene tree as well as the other five noncorresponding gene trees and the species tree. When comparing the maximum likelihood scores for these seven trees, the corresponding gene tree always gave the best likelihood scores (table 2). Trees most similar to the corresponding gene tree usually were better performers.
Maximum Likelihood Analysis
LRTs based on poorly fitting trees were more likely to be significant (table 2). In general, LRTs based on corresponding gene trees were the most conservative. Because corresponding gene trees provide the most rigorous examination of the data, they will be the focus of the remainder of the Results and Discussion.
LRT results were variable among cichlid opsin genes. All (M0–M3) rate heterogeneity LRTs were significant (P < 0.05), except for SWS2b (table 2). This indicates that relative rates of substitution are variable among sites in all opsin classes, except for SWS2b. Both (M1–M2) and (M7–M8) LRTs were significant (P < 0.05) for all remaining opsins, except SWS1. LRT results, ω estimates (table 3), and the prediction of sites that have evolved under positive selection (discussed below) suggest that a portion of sites in SWS2a, Rh2, LWS, and Rh1 cichlid opsin genes have evolved under positive selection.
Parameter Estimates of Nonsynonymous/Synonymous Substitution Rate Ratios Greater than One
. | . | . | . | Additional Class ω > 1 . | . | |
|---|---|---|---|---|---|---|
| Gene . | Model . | ω . | % Sites . | ω . | % Sites . | |
| SWS1 | M3 | 2.29 | 0.155 | |||
| SWS2a | M3 | 7.21 | 0.057 | |||
| M2 | 7.16 | 0.050 | ||||
| M8 | 3.74 | 0.141 | ||||
| Rh2 | M3 | 5.57 | 0.033 | 1.80 | 0.236 | |
| M2 | 2.66 | 0.229 | ||||
| M8 | 3.03 | 0.164 | ||||
| LWS | M3 | 9.61 | 0.029 | 1.91 | 0.186 | |
| M2 | 7.00 | 0.057 | ||||
| M8 | 3.72 | 0.162 | ||||
| Rh1 | M3 | 388.72 | 0.003 | 11.59 | 0.080 | |
| M2 | 17.54 | 0.055 | ||||
| M8 | 14.07 | 0.069 | ||||
. | . | . | . | Additional Class ω > 1 . | . | |
|---|---|---|---|---|---|---|
| Gene . | Model . | ω . | % Sites . | ω . | % Sites . | |
| SWS1 | M3 | 2.29 | 0.155 | |||
| SWS2a | M3 | 7.21 | 0.057 | |||
| M2 | 7.16 | 0.050 | ||||
| M8 | 3.74 | 0.141 | ||||
| Rh2 | M3 | 5.57 | 0.033 | 1.80 | 0.236 | |
| M2 | 2.66 | 0.229 | ||||
| M8 | 3.03 | 0.164 | ||||
| LWS | M3 | 9.61 | 0.029 | 1.91 | 0.186 | |
| M2 | 7.00 | 0.057 | ||||
| M8 | 3.72 | 0.162 | ||||
| Rh1 | M3 | 388.72 | 0.003 | 11.59 | 0.080 | |
| M2 | 17.54 | 0.055 | ||||
| M8 | 14.07 | 0.069 | ||||
Parameter Estimates of Nonsynonymous/Synonymous Substitution Rate Ratios Greater than One
. | . | . | . | Additional Class ω > 1 . | . | |
|---|---|---|---|---|---|---|
| Gene . | Model . | ω . | % Sites . | ω . | % Sites . | |
| SWS1 | M3 | 2.29 | 0.155 | |||
| SWS2a | M3 | 7.21 | 0.057 | |||
| M2 | 7.16 | 0.050 | ||||
| M8 | 3.74 | 0.141 | ||||
| Rh2 | M3 | 5.57 | 0.033 | 1.80 | 0.236 | |
| M2 | 2.66 | 0.229 | ||||
| M8 | 3.03 | 0.164 | ||||
| LWS | M3 | 9.61 | 0.029 | 1.91 | 0.186 | |
| M2 | 7.00 | 0.057 | ||||
| M8 | 3.72 | 0.162 | ||||
| Rh1 | M3 | 388.72 | 0.003 | 11.59 | 0.080 | |
| M2 | 17.54 | 0.055 | ||||
| M8 | 14.07 | 0.069 | ||||
. | . | . | . | Additional Class ω > 1 . | . | |
|---|---|---|---|---|---|---|
| Gene . | Model . | ω . | % Sites . | ω . | % Sites . | |
| SWS1 | M3 | 2.29 | 0.155 | |||
| SWS2a | M3 | 7.21 | 0.057 | |||
| M2 | 7.16 | 0.050 | ||||
| M8 | 3.74 | 0.141 | ||||
| Rh2 | M3 | 5.57 | 0.033 | 1.80 | 0.236 | |
| M2 | 2.66 | 0.229 | ||||
| M8 | 3.03 | 0.164 | ||||
| LWS | M3 | 9.61 | 0.029 | 1.91 | 0.186 | |
| M2 | 7.00 | 0.057 | ||||
| M8 | 3.72 | 0.162 | ||||
| Rh1 | M3 | 388.72 | 0.003 | 11.59 | 0.080 | |
| M2 | 17.54 | 0.055 | ||||
| M8 | 14.07 | 0.069 | ||||
(M3—Model B) branch-site variable LRTs detected significant differences (P < 0.05) in the relative rates of substitution between turbid and clear-water species for SWS1, Rh2, LWS, and Rh1 genes (table 4). When Lake Victoria (table 4) and Nile River (table 4) lineages were tested individually, the Lake Victoria lineage was never indicated to have a significantly different relative rate of substitution from the clear-water species, except for Rh1. In contrast, the Nile lineage was detected to be significantly different from clear-water lineages in all opsin genes except SWS2a and Rh1.
Branch-Site Variable LRT Results
Gene . | M3 . | MB . | LRT . | P value . | ||||
|---|---|---|---|---|---|---|---|---|
| Turbid: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,986.69 | −1,976.46 | 20.47 | 3.6E–05* | ||||
| SWS2a | −1,739.34 | −1,738.86 | 0.96 | 6.2E–01 | ||||
| SWS2b | −1,921.75 | −1,918.84 | 5.24 | 5.5E–02 | ||||
| Rh2 | −2,037.03 | −2,031.47 | 11.13 | 3.8E–03* | ||||
| LWS | −1,950.35 | −1,943.59 | 13.51 | 1.2E–03* | ||||
| Rh1 | −1,860.42 | −1,856.51 | 7.82 | 2.0E–02* | ||||
| Lake Victoria: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,780.77 | −1,779.16 | 3.22 | 2.0E–01 | ||||
| SWS2a | −1,655.83 | −1,655.14 | 1.37 | 5.0E–01 | ||||
| SWS2b | −1,754.82 | −1,753.84 | 1.95 | 3.8E–01 | ||||
| Rh2 | −1,889.40 | −1,889.38 | 0.02 | 9.9E–01 | ||||
| LWS | −1,813.14 | −1,813.14 | 0.00 | 1.0E+00 | ||||
| Rh1 | −1,754.15 | −1,746.95 | 14.40 | 7.5E–04* | ||||
| Nile River: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,945.88 | −1,934.63 | 22.52 | 1.3E–05* | ||||
| SWS2a | −1,700.99 | −1,700.99 | 0.00 | 1.0E+00 | ||||
| SWS2b | −1,880.49 | −1,877.43 | 6.12 | 4.7E–02* | ||||
| Rh2 | −2,011.34 | −2,003.04 | 16.58 | 2.5E–04* | ||||
| LWS | −1,895.04 | −1,887.88 | 14.31 | 7.8E–04* | ||||
| Rh1 | −1,771.39 | −1,770.30 | 2.18 | 3.4E–01 | ||||
Gene . | M3 . | MB . | LRT . | P value . | ||||
|---|---|---|---|---|---|---|---|---|
| Turbid: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,986.69 | −1,976.46 | 20.47 | 3.6E–05* | ||||
| SWS2a | −1,739.34 | −1,738.86 | 0.96 | 6.2E–01 | ||||
| SWS2b | −1,921.75 | −1,918.84 | 5.24 | 5.5E–02 | ||||
| Rh2 | −2,037.03 | −2,031.47 | 11.13 | 3.8E–03* | ||||
| LWS | −1,950.35 | −1,943.59 | 13.51 | 1.2E–03* | ||||
| Rh1 | −1,860.42 | −1,856.51 | 7.82 | 2.0E–02* | ||||
| Lake Victoria: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,780.77 | −1,779.16 | 3.22 | 2.0E–01 | ||||
| SWS2a | −1,655.83 | −1,655.14 | 1.37 | 5.0E–01 | ||||
| SWS2b | −1,754.82 | −1,753.84 | 1.95 | 3.8E–01 | ||||
| Rh2 | −1,889.40 | −1,889.38 | 0.02 | 9.9E–01 | ||||
| LWS | −1,813.14 | −1,813.14 | 0.00 | 1.0E+00 | ||||
| Rh1 | −1,754.15 | −1,746.95 | 14.40 | 7.5E–04* | ||||
| Nile River: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,945.88 | −1,934.63 | 22.52 | 1.3E–05* | ||||
| SWS2a | −1,700.99 | −1,700.99 | 0.00 | 1.0E+00 | ||||
| SWS2b | −1,880.49 | −1,877.43 | 6.12 | 4.7E–02* | ||||
| Rh2 | −2,011.34 | −2,003.04 | 16.58 | 2.5E–04* | ||||
| LWS | −1,895.04 | −1,887.88 | 14.31 | 7.8E–04* | ||||
| Rh1 | −1,771.39 | −1,770.30 | 2.18 | 3.4E–01 | ||||
NOTE.—“MB” refers to Model B. “*” indicates a significant LRT statistic (P ≤ 0.05 ; df = 2).
Branch-Site Variable LRT Results
Gene . | M3 . | MB . | LRT . | P value . | ||||
|---|---|---|---|---|---|---|---|---|
| Turbid: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,986.69 | −1,976.46 | 20.47 | 3.6E–05* | ||||
| SWS2a | −1,739.34 | −1,738.86 | 0.96 | 6.2E–01 | ||||
| SWS2b | −1,921.75 | −1,918.84 | 5.24 | 5.5E–02 | ||||
| Rh2 | −2,037.03 | −2,031.47 | 11.13 | 3.8E–03* | ||||
| LWS | −1,950.35 | −1,943.59 | 13.51 | 1.2E–03* | ||||
| Rh1 | −1,860.42 | −1,856.51 | 7.82 | 2.0E–02* | ||||
| Lake Victoria: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,780.77 | −1,779.16 | 3.22 | 2.0E–01 | ||||
| SWS2a | −1,655.83 | −1,655.14 | 1.37 | 5.0E–01 | ||||
| SWS2b | −1,754.82 | −1,753.84 | 1.95 | 3.8E–01 | ||||
| Rh2 | −1,889.40 | −1,889.38 | 0.02 | 9.9E–01 | ||||
| LWS | −1,813.14 | −1,813.14 | 0.00 | 1.0E+00 | ||||
| Rh1 | −1,754.15 | −1,746.95 | 14.40 | 7.5E–04* | ||||
| Nile River: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,945.88 | −1,934.63 | 22.52 | 1.3E–05* | ||||
| SWS2a | −1,700.99 | −1,700.99 | 0.00 | 1.0E+00 | ||||
| SWS2b | −1,880.49 | −1,877.43 | 6.12 | 4.7E–02* | ||||
| Rh2 | −2,011.34 | −2,003.04 | 16.58 | 2.5E–04* | ||||
| LWS | −1,895.04 | −1,887.88 | 14.31 | 7.8E–04* | ||||
| Rh1 | −1,771.39 | −1,770.30 | 2.18 | 3.4E–01 | ||||
Gene . | M3 . | MB . | LRT . | P value . | ||||
|---|---|---|---|---|---|---|---|---|
| Turbid: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,986.69 | −1,976.46 | 20.47 | 3.6E–05* | ||||
| SWS2a | −1,739.34 | −1,738.86 | 0.96 | 6.2E–01 | ||||
| SWS2b | −1,921.75 | −1,918.84 | 5.24 | 5.5E–02 | ||||
| Rh2 | −2,037.03 | −2,031.47 | 11.13 | 3.8E–03* | ||||
| LWS | −1,950.35 | −1,943.59 | 13.51 | 1.2E–03* | ||||
| Rh1 | −1,860.42 | −1,856.51 | 7.82 | 2.0E–02* | ||||
| Lake Victoria: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,780.77 | −1,779.16 | 3.22 | 2.0E–01 | ||||
| SWS2a | −1,655.83 | −1,655.14 | 1.37 | 5.0E–01 | ||||
| SWS2b | −1,754.82 | −1,753.84 | 1.95 | 3.8E–01 | ||||
| Rh2 | −1,889.40 | −1,889.38 | 0.02 | 9.9E–01 | ||||
| LWS | −1,813.14 | −1,813.14 | 0.00 | 1.0E+00 | ||||
| Rh1 | −1,754.15 | −1,746.95 | 14.40 | 7.5E–04* | ||||
| Nile River: clear branch-site variable LRT results | ||||||||
| SWS1 | −1,945.88 | −1,934.63 | 22.52 | 1.3E–05* | ||||
| SWS2a | −1,700.99 | −1,700.99 | 0.00 | 1.0E+00 | ||||
| SWS2b | −1,880.49 | −1,877.43 | 6.12 | 4.7E–02* | ||||
| Rh2 | −2,011.34 | −2,003.04 | 16.58 | 2.5E–04* | ||||
| LWS | −1,895.04 | −1,887.88 | 14.31 | 7.8E–04* | ||||
| Rh1 | −1,771.39 | −1,770.30 | 2.18 | 3.4E–01 | ||||
NOTE.—“MB” refers to Model B. “*” indicates a significant LRT statistic (P ≤ 0.05 ; df = 2).
Site Predictions
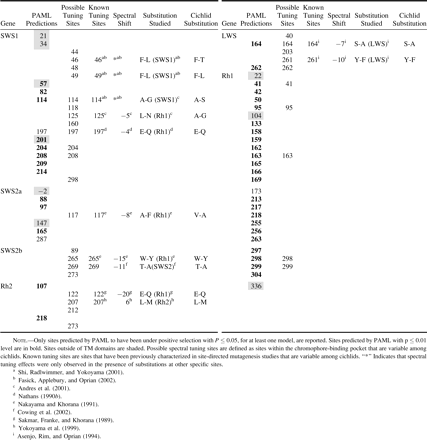
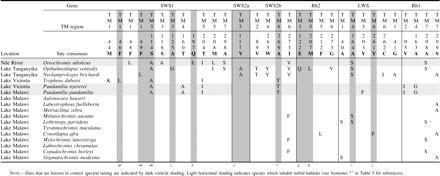
Based on the X-ray crystallographic studies of bovine rhodopsin, nearly all sites that modify the spectral properties of visual pigments are in the vicinity of/oriented toward the chromophore and make up the chromophore-binding pocket. Sites that are oriented toward the chromophore roughly comprise the set of possible tuning sites. Possible spectral tuning sites show transspecific variation in all opsin classes (table 5). The effects of substitution at many of these possible tuning sites have been characterized through mutagenesis studies (reviewed in Takahashi and Ebrey 2003; Yokoyama and Tada 2003). Those sites shown through mutagenesis studies to control opsin tuning comprise known tuning sites. With the exception of the Rh1 class, known tuning sites are transspecifically variable in all opsin classes (table 5).
PAML predicted amino acid sites (P ≤ 0.05) to be under positive selection for all opsin classes where LRTs were significant (table 5). We report the summation of PAML predictions from all models where sites are predicted. In the case of Rh2 and LWS M3, all nonsynonymous substitutions are predicted to be under positive selection. Because this is an unreasonable result, the predictions of these specific models are not included unless other models support them. Rh1 M8 predicts all but two nonsynonymous substitutions as having been under positive selection. We include these results because not all sites are predicted. Although most predictions (81%) fell within TM domains, a much smaller proportion (29%) of those site predictions are in the chromophore-binding pocket. A total of 31 possible spectral tuning sites are variable within the data. Further, many possible spectral tuning sites are not predicted to have been under positive selection (65%). Some sites within the chromophore-binding pocket have been previously characterized through mutagenesis studies and make up all known spectral tuning sites. A total of three known tuning sites are also PAML predictions. An additional eight known tuning sites are variable among cichlid opsins. Finally, PAML identifies eight possible tuning sites that have not been previously characterized by mutagenesis methods.
Discussion
This study examines the evidence for adaptive molecular evolution in cichlid opsin genes in response to gross differences in environmental light conditions. One might be tempted to conclude that the cichlid opsin gene with the least nucleotide divergence (SWS2a) has experienced the least divergent selection, relative to other opsin genes. However, nucleotide divergence alone is a poor comparative estimator of selective pressure as there are other factors that can be responsible for the observed variation in nucleotide sequence divergence, namely differences in the background rates of evolution, as determined by the rates of synonymous substitution. Variation, among genes, in the rates of synonymous substitution has been documented in previous studies (i.e., Senchina et al. 2003). We also observed variation in synonymous substitution rates among the cichlid opsins, further supporting previous findings that the relative selective regime is independent of the absolute number of nucleotide substitutions. Codon usage bias, GC content, and genomic location have all been proposed as possible causes of rate variation among genes (Wolf, Sharp, and Li 1989; Zhang, Vision, and Gaut 2002; Senchina et al. 2003). Interestingly, in cichlids, SWS2a, SWS2b, and LWS are located in a tandem array with 4.5 and 6 kb, respectively, of intervening sequence (Carleton and Kocher 2001). Given the close physical proximity of these three genes, it is of note that they still have very different rates of substitution, as can be extrapolated from the proportion of variable sites (SWS2a, 0.047; SWS2b, 0.068; and LWS, 0.078).
LRT Evidence for Positive Selection Among Cichlid Opsins
Site-specific LRTs indicate that amino acid sites in Rh2, LWS, and Rh1 have evolved at variable rates. The site-specific LRTs also show that these same opsins have experienced positive selection, a result that is supported by all model comparisons examined. Further, branch/site-specific LRTs indicate that relative rates of substitution are variable between turbid and clear-water species for Rh2 and LWS opsin genes. The failure of all LRTs indicate that SWS2a and SWS2b have evolved solely under purifying selection. The failure of selection and positive selection LRTs indicate that SWS1 has evolved under a regime of neutral evolution.
Variation Between Turbid and Clear lineages
Positive selection was detected among Rh2, LWS, and Rh1 genes using site-specific models. Branch/site-specific models identified Rh2, LWS, and Rh1 as having a class of sites with different relative rates of substitution between turbid and clear-water species (table 4). This suggests that longer wavelength absorbing genes are evolving under selective pressure induced by the photic environment. Branch/site-specific differences associated with water clarity are also seen in the SWS1 opsin, despite the failure of site-specific models to detect positive selection. This might indicate that some SWS1 opsins have evolved under neutral evolution, while others have been more restrained under purifying selection.
Examination of each turbid lineage separately yields only partially interpretable results (table 4). Because examination of individual turbid lineages required the removal of the other turbid lineage, the data available were also reduced. Reduction in data has been documented to reduce the power of the LRT (Anisimova, Bielawski, and Yang 2001). This reduction in power was most pronounced upon removal of the Nile lineage, which is among the most basal. The decrease in power caused by lineage removal may have decreased our ability to individually test the Lake Victoria lineage. This explains the lack of detectable difference between the Lake Victoria lineage and all the clear-water lineages (table 4). Further, Wong et al. (2004) note that in cases of directional selection where mutations rapidly reach fixation, the current methods may have difficulty. Data reduction did not appear to be a problem in the analysis of the Nile lineage (table 4) as the results show a similar pattern when the Lake Victoria and Nile River lineages are averaged.
Another reason we may not detect differences between Lake Victoria and clear lineages is that these tests average the selective regime over time. The signature of selection can be lost if there are multiple shifts in selective pressure over time. Several studies suggest that ancestors to the Lake Victoria lineage may have colonized other lakes, subsequently returning to the rivers before colonizing Lake Victoria (Nagl et al. 2000; Seehausen et al. 2003; Verheyen et al. 2003). If a previous colonization event or long-term shift in photic environment has occurred, this could potentially eliminate-diminish the difference in relative substitution rates among turbid Lake Victoria and present day clear-water lineages because of averaging of selective signature over time.
Evidence of Functional Divergence Among Cichlid Opsins
All opsin genes have substitutions that are known tuning sites, except Rh1 (table 5). Only the SWS1, SWSZb, Rh2, and LWS opsin classes have substitutions that are known to tune their respective classes. The transspecific variation at both possible and known spectral tuning sites demonstrates the potential for functional divergence in all opsin classes. PAML predicts that several known and possible tuning sites listed in table 4 may be under positive selection. Many of the sites predicted by PAML have not been characterized and therefore are particularly interesting candidates for study by site-directed mutagenesis.
If differences between turbid and clear environments have driven the evolution of East African cichlid opsin genes, one would expect that turbid lineages would have more long wavelength–shifting substitutions because turbid environments are long-wavelength shifted. One would also expect that turbid lineages would have unique sets of possible spectral tuning substitutions, relative to clear lineages. Independently evolved turbid lineages need not use the same spectral tuning substitutions; however, the substitutions should be unique relative to clear lineages. Because many of the substitutions observed are different from those previously studied or are at uncharacterized sites, the magnitude-directionality of spectral shifts caused by some substitutions cannot be predicted. Table 6 shows that only in SWS1 do turbid lineages have unique sets of possible spectral tuning substitutions (Nile lineage: 48, 114, 118, 197, 204, 208, and 298; Lake Victoria lineage: 114, 160, and 204). All but sites 48 and 160 were predicted by PAML to have been under positive selection. E197Q, a known short wavelength-shifting substitution, causes a −4 nm spectral shift. This is consistent with our expectations. Also, site 114 is known to be important in SWS1 tuning, although in mammals it acts in a synergistic manner in coordination with other sites that are not variable among these data (Shi, Radlwimmer, and Yokoyama 2001; Fasick, Applebury, and Oprian 2002). Among other opsin genes, there may be a lack of functional differentiation with respect to photic environment, or, alternatively, other sites that have not been considered as possible tuning sites might be responsible for functional differentiation. PAML highlights several uncharacterized possible tuning sites that may be import in spectral tuning (SWS1, 204 and 208; LWS, 262; Rh1, 41, 163, 298, and 299). These sites represent good candidates for site-directed mutagenesis studies, which will be needed to determine the effects and uniform directionality of spectral shifts (directional-positive selection) among lineages.
Possible Opsin Tuning Sites Based on Previous Functional and Structural Studies
 |
 |
Possible Opsin Tuning Sites Based on Previous Functional and Structural Studies
 |
 |
Conclusions
Given the remarkable ability of cichlids to utilize multiple nonexclusive mechanisms to tune visual sensitivity, the detection of variation in natural selection and the prediction of sites under positive selection are of note. Cichlids show variation in opsin gene expression, with different species expressing different subsets of cone opsins (Carleton and Kocher 2001). Further, variation in the molecular mechanisms of spectral tuning, both across sites and classes, may have an effect on the analysis. For example in bird SWS1, site 86 causes a 75-nm spectral shift (Shi, Radlwimmer, and Yokoyama 2001). Most known tuning sites, however, have a much smaller effect of less than 10 nm (reviewed in Yokoyama 2002; Takahashi and Ebrey 2003). Finally, species-specific ecological factors are also likely to play a role (Cummings and Partridge 2001). The present study focuses on gross differences in water clarity, although other factors such as depth are likely to be important in shaping visual sensitivity. A future analysis focusing on depth may therefore prove rewarding.
Unlike other molecules that have been the focus of molecular evolutionary computational studies, the clear link between opsin function and the environment, the availability of robust functional assays (i.e., spectral absorbance and transducin activation), and the rich body of mutagenic studies provide researchers with a well-characterized system to test molecular evolutionary models and specific ecological hypotheses. In this work, we demonstrated statistical evidence of positive selection in cichlid opsin genes. We then showed that there are differences in selective pressure among lineages that are known to have long-term residence in turbid habitats compared to lineages that inhabit clear photic environments. Finally, we identified candidate spectral tuning sites in cichlid opsin classes.
Photic environment–driven evolution may have played a significant role in the subsequent evolution of male nuptial hue usage and the diversity of color patterns for which cichlids are so renowned. Already researchers have noted that the color palette used by species living in turbid habitats is generally long-wavelength shifted (Seehausen 1999). This work begins to demonstrate a fundamental mechanism through which changes in hue usage are likely to be modulated.
Brian Golding, Associate Editor
This work has been supported by NSF grant IBN 0131285 to KLC. During the latter part of this work, TCS was supported by a UNH dissertation fellowship. Tissue collections were possible due to the Malawi government, the University of Malawi, the US Department of Education's Graduate Assistance in Areas of National Need Fellowship (to RCJ), the American Cichlid Association's Jordan Endowment (to RCJ), and NSF grant 9905127 (to TDK). The authors would like to thank Adriana Briscoe, Jay Stauffer, J. Todd Streelman, W. Kelley Thomas, and members of the Kocher lab for their assistance and thoughtful discussions. We also extend a special thanks to Ziheng Yang for technical assistance. Finally, we thank Nick Goldman, Brian Golding, and anonymous referrees for helpful comments.
References
Albertson, R. C., J. A. Markert, P. D. Danley, and T. D. Kocher.
Andres, A., A. Kosoy, P. Garriga, and J. Manyosa.
Anisimova, M., J. P. Bielawski, and Z. Yang.
Asenjo, A. B., J. Rim, and D. D. Oprian.
Bowmaker, J. K., V. I. Govardovskii, S. A. Shukolyukov, L. V. Zueva, D. M. Hunt, V. G. Sideleva, and O. G. Smirnova.
Briscoe, A. D.
Carleton, K. L., F. I. Harosi, and T. D. Kocher.
Carleton, K. L., and T. D. Kocher.
Civetta, A.
Cowing, J. A., S. Poopalasundaram, S. E. Wilkie, J. K. Bowmaker, and D. M. Hunt.
Crescitelli, F.
Cummings, M. E., and J. C. Partridge.
Ding, Y. C., H. C. Chi, D. L. Grady et al. (12 co-authors).
Fasick, J. I., M. L. Applebury, and D. D. Oprian.
Ford, M. J.
Graur, D., and W.-H. Li.
Hunt, D. M., J. Fitzgibbon, S. J. Slobodyanyuk, and J. K. Bowmaker.
Kocher, T. D., J. A. Conroy, K. R. McKaye, J. R. Stauffer, and S. F. Lockwood.
Lythgoe, J. N., W. R. A. Muntz, J. C. Partridge, J. Shand, and D. M. Williams.
Moran, P., and I. Kornfield.
Nagl, S., H. Tichy, W. E. Mayer, N. Takezaki, N. Takahata, and J. Klein.
Nakayama, T. A., and H. G. Khorana.
Nathans, J.
———.
Palczewski, K., T. Kumasaka, T. Hori et al. (12 co-authors).
Pamilo, P., and M. Nei.
Partridge, J. C., S. N. Archer, and J. N. Lythgoe.
Partridge, J. C., and M. E. Cummings.
Saitou, N., and M. Nei.
Sakmar, T. P., R. R. Franke, and H. G. Khorana.
Sakmar, T. P., S. T. Menon, E. P. Marin, and E. S. Awad.
Seehausen, O.
Seehausen, O., J. J. M. van Alphen, and F. Witte.
Seehausen, O., E. Koetsier, M. V. Schneider, L. J. Chapman, C. A. Chapman, M. E. Knight, G. F. Turner, J. J. van Alphen, and R. Bills.
Senchina, D. S., I. Alvarez, R. C. Cronn, B. Liu, J. Rong, R. D. Noyes, A. H. Paterson, R. A. Wing, T. A. Wilkins, and J. F. Wendel.
Shi, Y., F. B. Radlwimmer, and S. Yokoyama.
Streelman, J. T., R. Zardoya, A. Meyer, and S. A. Karl.
Sugawara, T., Y. Terai, and N. Okada.
Swanson, W. J., R. Nielsen, and Q. Yang.
Swofford, D. L.
Takahashi, Y., and T. G. Ebrey.
Teller, D. C., T. Okada, C. A. Behnke, K. Palczewski, and R. E. Stenkamp.
Terai, Y., W. E. Mayer, J. Klein, H. Tichy, and N. Okada.
Thorpe, A., R. H. Douglas, and R. J. Truscott.
van der Meer, H. J., and J. K. Bowmaker.
Verheyen, E., W. Salzburger, J. Snoeks, and A. Meyer.
Wolfe, K. H., P. M. Sharp, and W. H. Li.
Wong, W. S., Z. Yang, N. Goldman, and R. Nielsen.
Yang, Z.
———.
Yang, Z., and R. Nielsen.
Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen.
Yang, Z., and W. J. Swanson.
Yokoyama, S., and T. Tada.
Yokoyama, S., H. Zhang, F. B. Radlwimmer, and N. S. Blow.
Zhang, L., T. J. Vision, and B. S. Gaut.
Author notes
*Hubbard Center for Genome Studies and Department of Zoology, University of New Hampshire; †Institute of Zoology, Aquatic Ecology, University of Bern, Bern, Switzerland; ‡Department of Biomedical Sciences, Cornell University; and §Ecology, Evolution, and Natural Resources, Cook College, Rutgers University