-
PDF
- Split View
-
Views
-
Cite
Cite
Esther E. Schmidt, Oliver Pelz, Svetlana Buhlmann, Grainne Kerr, Thomas Horn, Michael Boutros, GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D1021–D1026, https://doi.org/10.1093/nar/gks1170
Close - Share Icon Share
Abstract
RNA interference (RNAi) represents a powerful method to systematically study loss-of-function phenotypes on a large scale with a wide variety of biological assays, constituting a rich source for the assignment of gene function. The GenomeRNAi database (http://www.genomernai.org) makes available RNAi phenotype data extracted from the literature for human and Drosophila. It also provides RNAi reagent information, along with an assessment as to their efficiency and specificity. This manuscript describes an update of the database previously featured in the NAR Database Issue. The new version has undergone a complete re-design of the user interface, providing an intuitive, flexible framework for additional functionalities. Screen information and gene-reagent-phenotype associations are now available for download. The integration with other resources has been improved by allowing in-links via GenomeRNAi screen IDs, or external gene or reagent identifiers. A distributed annotation system (DAS) server enables the visualization of the phenotypes and reagents in the context of a genome browser. We have added a page listing ‘frequent hitters’, i.e. genes that show a phenotype in many screens, which might guide on-going RNAi studies. Structured annotation guidelines have been established to facilitate consistent curation, and a submission template for direct submission by data producers is available for download.
INTRODUCTION
Double-stranded RNAs have been shown to trigger the degradation of homologous mRNA across a wide spectrum of species, a mechanism termed RNA interference (RNAi) (1–3). RNAi has become a very powerful experimental method used to systematically silence gene expression on a large scale. High-throughput RNAi screening experiments allow the determination of loss-of-function phenotypes in a wide variety of biological assays and therefore represent an important approach in the assignment of gene function.
A growing amount of RNAi screening data for various species has become available in the literature, and the collection and integration of these data represents a major challenge. The urgent need for a public repository for RNAi screening data has recently been emphasized (4). To make better use of the wealth of RNAi screening data, it is also essential to be able to compare data from different experiments. This demands a standardization of the data representation, which constitutes a formidable challenge, given the vast variety of assays performed. In recent releases of GenomeRNAi, we have attempted to address this issue by the definition of structured annotation guidelines, using controlled vocabularies wherever possible.
The GenomeRNAi database (http://www.genomernai.org) has been described in two previous NAR database issues (5,6). The 2010 version contained 97 screens from Drosophila and 48 screens in human cells as well as ∼100 000 RNAi reagents for each species. Here, we describe an updated version of the GenomeRNAi database with major additions and improvements. The user interface has undergone a complete re-design, creating an intuitive, user-friendly website. The new version of GenomeRNAi contains 170 screens performed in Drosophila, and 127 screens in human cells. New functionalities include download options, a ‘frequent hitters’ page, and providing the data via a DAS server. GenomeRNAi screen identifiers have been introduced, and in-links via common external gene identifiers have been enabled. A data submission template, based on the newly established structured annotation guidelines, has been made available.
DATABASE CONTENT
The GenomeRNAi database is populated by manual curation of RNAi screening data from literature. In general, the type of data collected can be divided into two categories: information about the screen itself and gene-reagent-phenotype associations. When a publication contains experiments performed in e.g. different cell lines, or with different assays, or when it includes follow-up screens, these are recorded as individual screens. As of release 9 (September 2012) the GenomeRNAi database contains 127 screens in human, and 170 in Drosophila, 53 of which have been performed in vivo. The number of phenotype entries has increased to more than 500 000.
GenomeRNAi also holds information about RNAi reagents, such as sequence, primer details, as well as information on the source RNAi reagent library. Currently, the database contains more than 460 000 reagents from 14 RNAi libraries. Reagents, for which sequence information is available, are regularly assessed as to their specificity and efficiency, and are re-mapped to the current genomic sequence using the NEXT-RNAi software (7).
Annotation guidelines
To allow comparison of RNAi screening data within the database, the data must be curated in a consistent, standardized manner. Given the large variety of assays and methods employed across different experiments and publications, such standardization represents a major challenge. To address this task, we have developed annotation guidelines, to be followed by curators. Wherever possible, we have defined a controlled vocabulary (CV) of pre-defined terms or, when the number of possible options was prohibitive, established vocabulary guidelines (VG). For example, the data field ‘biosource’ can have only one of (currently) four options out of a CV, namely ‘cell line’, ‘primary cells’, ‘tissue’ or ‘organism’. For the data field ‘assay’, the entries should follow examples given in the VG, e.g. ‘<cell type> morphology’, ‘<gene name> protein expression’, ‘viability’, ‘viability (synthetic lethal)’, etc. The annotation guidelines provide instructions for each data field, constituting a structured framework for consistent data curation. All RNAi screening data in GenomeRNAi that have been published in 2010 or later have been entered according to these annotation guidelines. For older screens not yet updated to the new standard (currently representing about one third of the total), an ongoing process of re-curation is in place.
Mapping to Entrez Gene ID
Another challenge faced when curating RNAi screening data from the literature is the variety of gene and reagent identifiers used by the authors. As a result, data from different sources cannot be directly compared. Therefore, we attempt to undertake a mapping from author-provided identifiers or symbols to the Entrez Gene ID, which we use as the reference identifier for GenomeRNAi. This enables the presentation on one page and the comparison of all data relating to a given Entrez Gene ID.
GenomeRNAi screen IDs
We have implemented screen identifiers for all screens in GenomeRNAi to allow unambiguous identification and access from external web resources via in-links. GenomeRNAi screen IDs follow the pattern ‘GRxxxxx-A(-x)’ with ‘xxxxx’ being a running number, and ‘-x’ being an optional suffix to indicate the running number of multiple screens published within a single publication. Screen IDs ending on ‘-0’ indicate that the screen in question has not been updated yet according to the new annotation guidelines. The ‘-0’ is removed upon re-curation, or replaced by ‘-1’, ‘-2’, etc. in the case of multiple screens in the publication. The ‘core’ identifier ‘GRxxxxx-A’ remains stable.
WEBSITE FEATURES
In 2011, the GenomeRNAi website underwent a complete re-design. A user-friendly web interface was created and a framework for the display of nested data features set in place.
Entry points and search options
The home page gives general information and latest news about the GenomeRNAi project. It further serves as entry point for the search functionality, allowing the user to search the database by gene symbol or identifier, by reagent identifier or by phenotype. It also provides a browse option, linking to a tabular list of all screens in GenomeRNAi, as well as a number of examples directly linked to different types of search result pages. Menu buttons at the top of the page direct the user e.g. to the download page, a site index or help pages (see Supplementary Data). At the bottom of the page, the user can sign up for the GenomeRNAi newsletter, or follow GenomeRNAi on Facebook or Twitter.
Data output
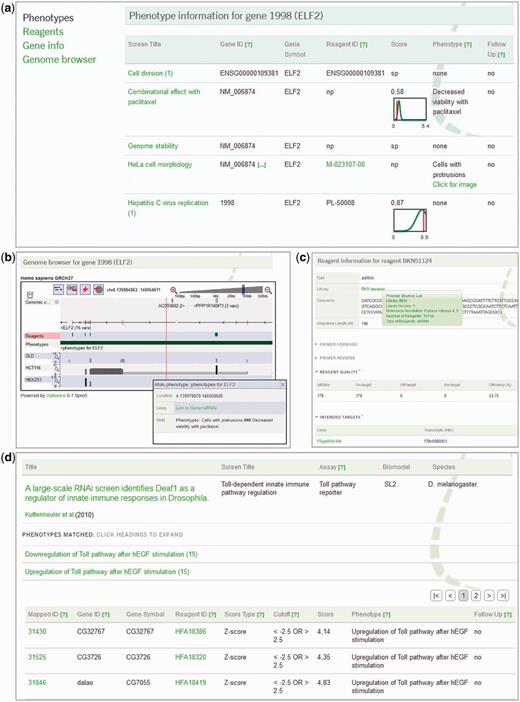
Some examples of data output pages are given in Figure 1. When searching for a gene or a reagent, the output is organized into four sections that can be selected by tabs in the upper left corner. On the gene details page, the first tab ‘Phenotypes’ (default) lists all phenotypes that have been identified for this gene in different screens (Figure 1a). The table contains a short screen title, hyperlinked to more details about the screen, as well as author-provided gene and reagent identifiers, and the score and phenotype recorded for this gene. For numerical scores, a graphical display of the score distribution and the position of the respective score are shown. If reagent details are available in the database, the reagent identifier is hyperlinked to the corresponding page. The second tab opens a list of reagents that have been reported to target the gene in question. Here, more information about the reagents and the corresponding reagent libraries are provided via hyperlinks. The third tab concerns the gene itself, making available details like chromosomal location, alternative names, links to external resources, as well as homology information between human and Drosophila. Finally, the fourth tab offers a dynamic genome browser display of the reagents and phenotypes in the genomic context, along with RNASeq data (unpublished data) from human or Drosophila cell lines, respectively, allowing the user to assess gene expression in the corresponding region (Figure 1b). For a reagent search, the data output is equivalent, with the first (default) tab providing reagent details such as sequence, primer characteristics, quality assessment and gene target information as generated by NEXT-RNAi (7) (Figure 1c).
Examples of data output pages. (a) Gene details page for the human gene ELF2, Entrez Gene ID 1998. Information on a gene is arranged into four tabs; the ‘Phenotypes’ tab opens by default. A list of screens that included the respective gene is displayed, along with author-provided gene IDs, gene symbols and reagent identifiers, as well as the scores and phenotypes recorded. For numerical scores, a little image underneath indicates the distribution of the scores and the relative position of the score in question. Screen titles are hyperlinked and display more details on the screen when clicked upon. The screen ‘Combinatorial effect with paclitaxel’ recorded the phenotype ‘Decreased viability with paclitaxel’ for the ELF2 gene. For the screen ‘HeLa cell morphology’, the phenotype is given as ‘Cells with protrusions’, and for this screen the user can open an image for direct assessment of the phenotype. (b) Dynamic genome browser display for the human ELF2 gene (fourth tab). RNAi reagents and phenotypes are displayed via the DAS technology in a Dalliance browser (8). RNASeq data for three human cell lines are provided as additional tracks at the bottom. Clicking on the phenotype track opens a window with information on the genomic location, the phenotypes recorded for this gene and a link to the respective gene details page in GenomeRNAi. The user can modify the display by zooming and scrolling, and also by adding additional tracks for data sources available from the DAS registry. (c) Reagent details page for the reagent BKN51124, targeting the cdc2 gene in Drosophila. The type of reagent, source library information and the sequence of the reagent are shown. Clicking on ‘details’ next to the library name, a mouse-over window displays additional information on the source library. The library name is hyperlinked to the library provider’s website. Additional sections provide primer characteristics, a quality assessment obtained by the NEXT-RNAi software (7) as well as information on the intended target gene for the selected reagent. If applicable, off-target genes are also displayed. (d) Screen-phenotype display for a Toll pathway screen by Kuttenkeuler et al. The screen has been selected on the ‘Browse’ page, followed by clicking on the ‘View Phenotypes’ button. Some key details on the screen are shown at the top row, including the publication title, hyperlinked for additional screen information, then a short screen title, as well as details on the assay, the biomodel and the species used in the experiment. This is followed by a list of phenotypes identified in the selected screen, along with the number of entries associated with each phenotype. Upon clicking on a phenotype, a table of genes recorded as showing this phenotype opens up. This table provides a hyperlinked Entrez Gene ID for all entries that could successfully be mapped. Furthermore, it contains author-provided gene and reagent details, followed by information on score type, applied score cut-off and the score itself. The ‘Follow Up’ column indicates whether further experiments have been performed with the aim of confirming the phenotype.
The user can also browse through all screens included in Genome RNAi. The screens are provided as a list that opens up nested sections, with screen details appearing upon clicking on a particular screen title. This nested section displays the abstract, screen details such as biomodel, assay and score type, as well as function buttons allowing the user to view all phenotypes identified in the screen, download the screen data, or connect to the respective PubMed entry. The ‘View Phenotypes’ button links to a page listing all distinct phenotypes described in the screen along with the number of times it has been identified, which in turn provides a nested list of all genes/reagents that have actually shown this phenotype (Figure 1d). At the bottom of the ‘Browse’ page, there is a text box that allows the user to filter the screen list by search terms, e.g. when looking for a particular author.
Download options
All screening data in GenomeRNAi are now available for download, either on a per screen basis via a download button on an individual screen details page or as bulk download from the download page. For the bulk download, one file is provided per species. The download format is explained on the download page. Briefly, the downloadable flat file is divided into two sections, the first one providing information about the screen itself, such as publication details, assay, biomodel and score type, the second one containing the gene-phenotype associations and reagent information in tab-delimited format. These data are provided via a ‘Creative Commons Attribution 3.0 Unported License’, i.e. users are free to make use of the data as they wish as long as appropriate credit is given.
Submission of data
Direct data submission by data producers is encouraged, and we make available a submission template for download. This Excel file contains instructions and examples for all data fields, following the annotation guidelines. Currently, the submission process involves communication with the author by e-mail, but the implementation of a login space for authors, allowing them and reviewers to view the data on the GenomeRNAi website, is underway.
Frequent hitters
As of release 8.0 the GenomeRNAi website features ‘frequent hitters’ lists. For a given species, these lists show genes in descending order with respect to the number of times they have shown a phenotype. All genes with at least two hits (= positive phenotypes) are listed, along with the RNAi reagents used. If an identical phenotype has been reported for the same gene in subscreens of the same publication, the phenotype has been counted only once. These frequent hitter lists can be used as a quick reference to identify genes that might play a role in multiple biological processes and therefore represent central network nodes, or genes the perturbation of which may be associated to off-target effects elicited by particular RNAi reagents. A download option is provided.
INTEGRATION WITH EXTERNAL RESOURCES
Integration with external resources is an essential requirement for a database to be useful to the scientific community. We therefore aim to enable the user to seamlessly follow links to relevant information in other resources to obtain a comprehensive view on the wider biological context of the data in question.
External links
The GenomeRNAi website is well inter-linked with other publically available databases. Genes in GenomeRNAi are linked to Ensembl (9), FlyBase (10), GeneCards (11), HGNC (12), HPRD (13), miRBase (14), OMIM (15), RefSeq (16), UniProt (17) and Vega (18). Reagents have links to the websites of the respective library providers.
In-links
Links to GenomeRNAi have been implemented on the FlyBase and UniProt websites and are scheduled to be included in the November 2012 release of the GeneCards website. GenomeRNAi data are also available as a data source in FlyMine (19), a popular data mining tool with fly biologists.
DAS server
As a further tool to integrate GenomeRNAi data with other resources, information is hosted on a Distributed Annotation System (DAS) server (20), providing reagent and phenotype information in their genomic context for both human and Drosophila. The GenomeRNAi DAS server is listed in the DAS registry (21) or can be accessed directly under http://genomernai.de/DASGenomeRNAi/das/.
CONCLUSION AND OUTLOOK
The GenomeRNAi database provides the largest publically available resource for RNAi reagents and phenotypes. The database currently hosts RNAi screening data from human and Drosophila but there are no technical restrictions to extending the scope to other species. We aim for comprehensive coverage of all large-scale RNAi experiments described in the literature, while at the same time encouraging direct submissions by data producers.
Standardized, consistent data representation remains a challenge. To some degree, this is being addressed by the ‘Minimum Information About an RNAi Experiment’ (MIARE) effort (http://miare.sourceforge.net/) though this is more concerned with the description of the experiment itself and less with the description of the resulting phenotypes. For GenomeRNAi curation, we have developed structured annotation guidelines, consistent with MIARE where applicable. More efforts will be required from the RNAi screening community toward the development of a generally accepted ontology for phenotype annotation. Equally important will be the support of journal editors in requesting authors to deposit their data in a structured format into a data repository when publishing their RNAi screening data.
In addition to working toward better coverage of RNAi data in GenomeRNAi, we aim to develop better resources for scientists to make use of the data. Comparison of screens is an important aspect and we are currently working on the implementation of a graphical screen comparison tool. Advanced search and filter options will allow the user to select a list of genes/screens to be displayed in this tool, or to be passed on to external functional analysis tools, such as STRING (22), or DAVID (23).
Based on the essentially unbiased nature of large-scale RNAi screens and the increasing amount of data obtained from such screens, we expect meta-analyses of this wealth of information to become progressively important. With the recent and prospective improvements of GenomeRNAi, we not only aim to make available RNAi screening data in standardized formats but to provide a useful tool to conduct meta-analyses and explore as yet unknown functional relationships between genes, pathways and phenotypes.
FUNDING
European Community’s Seventh Framework Programme [FP7/2007-2013] (CancerPathways); Helmholtz Alliance for Systems Biology. Funding for open access charge: German Cancer Research Center (DKFZ).
Conflict of interest statement: None declared.
ACKNOWLEDGEMENTS
The authors thank Klaus Yserentant, Arunraj Dhamodaran, Maximilian Koch and Andreas Kling for excellent support. They are grateful to Marco Breinig for critical comments on the manuscript.
REFERENCES
Author notes
Present address: Thomas Horn, Novartis Institutes for Biomedical Research, 250 Massachusetts Avenue, Cambridge, MA 02139, USA.




Comments