-
PDF
- Split View
-
Views
-
Cite
Cite
Aron Marchler-Bauer, Chanjuan Zheng, Farideh Chitsaz, Myra K. Derbyshire, Lewis Y. Geer, Renata C. Geer, Noreen R. Gonzales, Marc Gwadz, David I. Hurwitz, Christopher J. Lanczycki, Fu Lu, Shennan Lu, Gabriele H. Marchler, James S. Song, Narmada Thanki, Roxanne A. Yamashita, Dachuan Zhang, Stephen H. Bryant, CDD: conserved domains and protein three-dimensional structure, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D348–D352, https://doi.org/10.1093/nar/gks1243
Close - Share Icon Share
Abstract
CDD, the Conserved Domain Database, is part of NCBI’s Entrez query and retrieval system and is also accessible via http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml. CDD provides annotation of protein sequences with the location of conserved domain footprints and functional sites inferred from these footprints. Pre-computed annotation is available via Entrez, and interactive search services accept single protein or nucleotide queries, as well as batch submissions of protein query sequences, utilizing RPS-BLAST to rapidly identify putative matches. CDD incorporates several protein domain and full-length protein model collections, and maintains an active curation effort that aims at providing fine grained classifications for major and well-characterized protein domain families, as supported by available protein three-dimensional (3D) structure and the published literature. To this date, the majority of protein 3D structures are represented by models tracked by CDD, and CDD curators are characterizing novel families that emerge from protein structure determination efforts.
INTRODUCTION
Protein domains were initially described as stable or autonomously folding units of protein structure, inspired by first results from the experimental characterization of protein three-dimensional (3D) structure. This definition of protein domains tends to coincide remarkably often with what has emerged from systematic analyses of sequence data—the characterization of protein domains as units of molecular evolution. The majority of protein domain models collected in databases such as Pfam (1) and SMART (2), the contents of which have been incorporated into Conserved Domain Database (CDD), stem from the results of such sequence analyses, and the CDD in-house curation effort has adopted a similar view of protein domains. Even if the analysis of 3D structure suggests the presence of two or more structurally autonomous units, or ancient rearrangements at the gene level have resulted in tandemly repeated units, a domain model would not be split into smaller parts unless the analysis of sequence and structure databases strongly suggests that fragments homologous to one or more such smaller parts exist in different contexts.
While domain models encountered in CDD may not always reflect domains as inferred from analysis of 3D structure, the CDD curation effort does make use of 3D structure to delineate the boundaries of domain footprints and to guide MSA (multiple sequence alignment) models so that they agree with the results of 3D structure superposition. Protein 3D structure also serves as the template to define an MSA’s core block structure, following a simple model where structurally conserved and/or buried elements of protein 3D structure correspond to sequence regions that do not accumulate length variation in molecular evolution. In the CDD alignment model, all length variation is represented by unaligned regions between such conserved blocks, and unaligned regions often correspond to loops between secondary structure elements in protein 3D structure. Although this may seem oversimplified, it has not hampered our ability to utilize such MSAs for the detection of evolutionarily conserved and functionally distinct sub-families, which are often curated as separate models in hierarchically organized representations of domain superfamilies. CDD’s hierarchical classifications are cross-validated by CDD curators against classifications available in the published literature, and more recently against computationally generated classifications based on the detection of signature sequence motifs characteristic for protein domain families and sub-families (3).
Protein 3D structure also serves as evidence for functional sites that are annotated on conserved domain models, such as active sites and binding sites for substrates, cofactors, drugs, nucleic acids and other polypeptides. As CDD employs 3D structure throughout the model building and annotation process, the curation effort has benefited greatly from the increase in available 3D structure data as brought on by the Protein Structure Initiative (4) and the continuous growth of the Protein Data Bank (5).
The current version of CDD, v3.08, contains 43 212 alignment models, of which 8566 have been curated by NCBI. Other models originate from Pfam (1), SMART (2), COG (6), TIGRFAMs (7) and the NCBI Protein Clusters database (8). For detailed explanations of the tools and data provided by CDD, we direct the reader to the on-line documentation and earlier manuscripts (9). Here we give a brief account of recent changes to the protein sequence annotation services and discuss mutual coverage between conserved domain models and protein 3D structures.
CDD’S COVERAGE OF PROTEIN SEQUENCES AND 3D STRUCTURES
Currently, about 76% of protein sequences in Entrez (excluding sequences from metagenomes, which have been obtained via environmental sampling and are not included in the pre-computed annotation set) can be matched to one or more conserved domain models, inferring the function of more than 39 million publicly available proteins. When looking at only the subset of structure-linked protein sequences, which are derived from the processing of 3D structure (currently 213 507 entries), the fraction goes up to 94%, and is close to 98% when only considering structure-linked proteins with 50 amino acid residues or more (currently 199 602 entries). Somewhat higher numbers were recently reported by Xu and Dunbrack, using a multi-level procedure that combines sensitive profile searches with comparisons of protein 3D structure (10). After removing redundant data, we find around 1000 sequences derived from protein structure records that might belong to previously uncharacterized protein domain families. We have recently set up a monitoring system to track such 3D structure-derived proteins that are not covered by any domain model. From that list, we pick candidates for the curation of new domain models, particularly those that have related sequences, as identified by protein BLAST, with a wide phylogenetic spread. A large fraction of the cases turn out to be distant members of domain families that are already represented in CDD, at which point we schedule the family for re-curation so that we can extend its scope. Others are used as seeds to build new models.
CDD currently contains 94 domain superfamilies that are made up entirely of NCBI-curated domain models, and 23 of them contain more than one model in a hierarchical arrangement. Many of these were created as the result of surveying 3D structures without coverage, and after 1½ years of tracking coverage, we have seen the fraction of structure-derived proteins that are 50 residues long or longer and not covered by any domain model drop from 3% to about 2% (these figures also include structures that are not being considered for instantiating conserved domain models, including those where processing of 3D structures does not identify actual protein sequences and de-novo/designed proteins).
DOMAIN SUPERFAMILIES AND AVAILABILITY OF 3D STRUCTURES
A protein domain superfamily can be thought of as a set of protein sequence fragments that are related by common descent. In CDD, many such superfamilies are represented by a single-domain model, whereas others may be represented by a large number of models. In order to simplify sequence annotation displays, CDD clusters single-domain models that provide overlapping and partially redundant annotation into representations of protein domain superfamilies, which get assigned their own accessions with the prefix ‘cl'. Alignment models that appear to cover more than one single-domain footprint are flagged as multi-domain models and excluded from the clustering. Single-linkage clustering is performed on the remaining single-domain models, utilizing the pre-computed sequence annotation data for all sequences in the Entrez protein database (excluding sequences from metagenomes, which are currently not neighbored). Criteria for clustering are overlapping annotation intervals on sets of sequences with sufficient diversity, after applying conservative thresholds for RPS-BLAST E-values and overlapping interval size. The thresholds have been adjusted over time as the CDD and the protein sequence databases have grown significantly. More recently, CDD also maintains a curated list of prohibited linkages, to avoid false clustering, which may be triggered by problems with alignment model, protein sequence data and the neighboring method. Superfamily clusters are assigned accessions starting with ‘cl’, and clusters with more than one constituent alignment model are indexed for searching in Entrez, currently a total of 3295. The majority of the conserved domain superfamilies (9012) are represented by a single alignment model. The largest superfamily cluster at this point unites more than 500 single-domain models (cl09099, the P-loop_NTPase Superfamily).
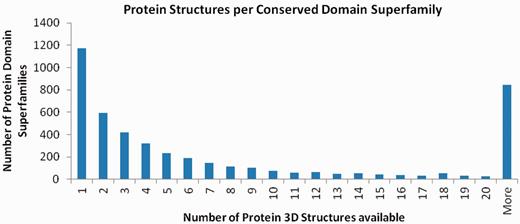
In the current version of CDD (on 1 October 2012) 5007 out of 12 307 single-domain superfamilies are linked to one or more 3D structures, suggesting that 3D structures are known for at least 41% of protein domain superfamilies. Almost one quarter of these 5007 superfamilies are represented by only a single 3D structure as available in NCBI’s Molecular Modeling Database (MMDB) (11). Figure 1 illustrates the distribution of domain superfamilies across available structure counts. Redundancy in the 3D structure data set does shift the distribution toward higher counts, of course. The 7300 superfamilies without a single representative structure are not shown.
This histogram illustrates the distribution of protein 3D structures between conserved domain superfamilies. Although the majority of superfamilies cannot be linked to a 3D structure representative, about one quarter of those that can be linked have only a single representative 3D structure. Data prepared with NCBI FLink (http://www.ncbi.nlm.nih.gov/Structure/flink/flink.cgi).
Of all proteins in NCBI’s Entrez database (excluding sequences from metagenomes), about 51% can be related to a known 3D structure via protein-BLAST searches. When establishing relationships via conserved domain models, that number goes up to over 60% (as estimated from a random sample of domain models), perhaps demonstrating the higher sensitivity of sequence-profile searches versus direct sequence comparison.
SPECIFIC HITS AND HIGH-CONFIDENCE ANNOTATION
CDD uses a simple mechanism to assign high confidence to matches between protein query sequences and domain models. In order to qualify as a ‘specific hit’, a match must be the highest-ranked match for the respective region of the query, and it must also cross a model-specific score threshold. The latter is determined automatically using the sequences that were employed in piling up the model’s MSA. The lowest scoring sequence from the model determines the model’s score threshold (12). Previously, this has only been applied to models curated by the NCBI CDD team, but recently we have started to use the same mechanism for all the models in the collection, and the most recent version of the CD-Search (13) service represents that change in how it presents concise and detailed search results. High-confidence domain annotation is now returned as provided by imported models, such as those from Pfam, and is particularly helpful when no NCBI-curated model is available. Therefore, the specific hits section of CD-Search results now displays the highest-scoring models from the NCBI curation effort, or in their absence, the highest-scoring models imported from external databases that meet the domain-specific score threshold.
QUERYING CDD WITH NUCLEOTIDE SEQUENCES
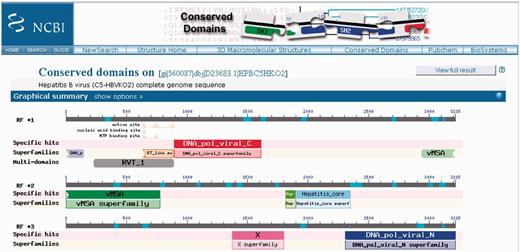
The CD-Search service now also accepts nucleotide sequences as queries. Nucleotide queries will be translated in all six reading frames, and corresponding polypeptide sequences will be searched against the model database. The results will be summarized so that the reading frames that pick up conserved domain hits are identified. Figure 2 gives an example of a CD-Search graphical results summary obtained for a nucleotide query representing a complete viral genome.
CD-Search results for a nucleotide query sequence, the complete genome sequence of a Hepatitis B virus. Results have been obtained for three different reading frames used for translation of the nucleotide query. Consequently, the display is split into three panels, which are labeled with ‘RF +1’, ‘RF +2’ and ‘RF +3’.
FUNCTIONAL SITE ANNOTATION
Conserved Domain models curated by NCBI often carry annotation of functional sites. These are recorded as co-ordinates on the MSA and resulting position-specific score matrices, and are mapped to protein query sequences via the CD-Search service. Site annotation is also pre-computed for proteins in Entrez and is readily available via the Entrez protein GenPept views or the graphical protein sequence viewer. To date, 18 263 sites have been recorded on 7382 models (about 86% of all NCBI-curated conserved domain alignments). Recently, we have increased the specificity of functional site mapping by adding sequence motifs/patterns to the site definitions, so that mapping is only performed when (i) a large fraction of the site’s positions can be mapped onto the protein query via the RPS-BLAST alignment; and (ii) the corresponding amino acid residues on the query sequence match the site motif, if present. Although this should reduce the number of erroneous site assignments, it allows the curation staff to annotate functional sites that might not be conserved across a more diverse set of domain family members, such as post-translational modification sites. We have compared CDD-based site annotations with those available in SwissProt, and while there is considerable overlap, specifically for sites categorized as active sites, the two sources of annotation complement each other to some degree (14).
UTILIZING CDD ANNOTATION DATA
In Table 1 we summarize access points to CDD and the CDD-based annotation data. Sequence annotation with conserved domain footprints and derived functional sites can be obtained via the CD-Search service, or via BATCH CD-Search for larger sets of query sequences. Domain architectures can be analyzed via the Conserved Domain Architecture Retrieval Tool (CDART) service (15), which has been completely overhauled recently. Both CDART’s performance and user interface have been improved. CDART enables users to search for proteins that have domain architectures similar to that of the query, and links to CDART are provided on CD-Search annotation displays. Domain architectures retrieved by CDART are ranked by similarity to the query’s domain architecture and by the architectures’ number of unique sequences.
URLs and other resources associated with the CDD project
| CDD | Database home page | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
| CDD help | CDD help documentation | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml |
| CDD FTP | CD models and alignments, pre-built search databases | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd |
| CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| Batch CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi |
| CDART | Domain architecture viewer | http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi |
| CDART FTP | Data summarizing conserved domain architectures | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdart/ |
| CDTree/Cn3D | Domain hierarchy viewer and editor | http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml |
| RPS-BLAST | Stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution | ftp://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
| CDD | Database home page | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
| CDD help | CDD help documentation | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml |
| CDD FTP | CD models and alignments, pre-built search databases | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd |
| CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| Batch CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi |
| CDART | Domain architecture viewer | http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi |
| CDART FTP | Data summarizing conserved domain architectures | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdart/ |
| CDTree/Cn3D | Domain hierarchy viewer and editor | http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml |
| RPS-BLAST | Stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution | ftp://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
URLs and other resources associated with the CDD project
| CDD | Database home page | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
| CDD help | CDD help documentation | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml |
| CDD FTP | CD models and alignments, pre-built search databases | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd |
| CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| Batch CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi |
| CDART | Domain architecture viewer | http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi |
| CDART FTP | Data summarizing conserved domain architectures | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdart/ |
| CDTree/Cn3D | Domain hierarchy viewer and editor | http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml |
| RPS-BLAST | Stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution | ftp://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
| CDD | Database home page | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
| CDD help | CDD help documentation | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml |
| CDD FTP | CD models and alignments, pre-built search databases | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd |
| CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| Batch CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi |
| CDART | Domain architecture viewer | http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi |
| CDART FTP | Data summarizing conserved domain architectures | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdart/ |
| CDTree/Cn3D | Domain hierarchy viewer and editor | http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml |
| RPS-BLAST | Stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution | ftp://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
FUNDING
Funding for open access charge: Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Paul Thiessen, Lianyi Han and the NCBI Information Engineering Branch for assistance with software development. We are grateful to the authors of Pfam, SMART, COGs, TIGRFAMs and NCBI’s Protein Clusters database for providing such vital resources.




Comments