-
PDF
- Split View
-
Views
-
Cite
Cite
Kyle R. Conway, Christopher N. Boddy, ClusterMine360: a database of microbial PKS/NRPS biosynthesis, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D402–D407, https://doi.org/10.1093/nar/gks993
Close - Share Icon Share
Abstract
ClusterMine360 (http://www.clustermine360.ca/) is a database of microbial polyketide and non-ribosomal peptide gene clusters. It takes advantage of crowd-sourcing by allowing members of the community to make contributions while automation is used to help achieve high data consistency and quality. The database currently has >200 gene clusters from >185 compound families. It also features a unique sequence repository containing >10 000 polyketide synthase/non-ribosomal peptide synthetase domains. The sequences are filterable and downloadable as individual or multiple sequence FASTA files. We are confident that this database will be a useful resource for members of the polyketide synthases/non-ribosomal peptide synthetases research community, enabling them to keep up with the growing number of sequenced gene clusters and rapidly mine these clusters for functional information.
INTRODUCTION
The amount of information on microbial secondary metabolite biosynthesis has been growing explosively. Gene clusters responsible for the biosynthesis of polyketides and non-ribosomal peptides, identified by the presence of polyketide synthases (PKS) or non-ribosomal peptide synthetases (NRPS) encoding genes, have received significant attention, resulting in the sequencing of hundreds of gene clusters. With the power, speed and low cost of next-generation sequencing methods, this number is expected to rapidly increase by at least an order of magnitude in the next few years.
To take advantage of this wealth of data, it needs to be easily accessible and discoverable. Although the sequences themselves are available in National Center for Biotechnology Information (NCBI) databases (1,2), they are frequently difficult to locate, partially because of the large amounts of information that these databases host. There is no standardized annotation for these biosynthetic gene clusters. For example, some are tagged with PKS and/or NRPS, such as the cycloheximide (accession number JX014302; Shen,B. and Yin,M., unpublished data) and streptothricin (accession number AB684619; Maruyama,C., Toyoda,J., Kato,Y., Izumikawa,M., Takagi,M., Shinya,K., Katano,H., Utagawa,T. and Hamano,Y., unpublished data) gene clusters, whereas others are tagged with the term polyketide synthase or non-ribosomal peptide synthetase, such as laidlomycin (accession number JQ793783; Hwang,J.Y., Kim,H.S., Sedai,B. and Nam,D.H., unpublished data) and collismycin A (accession number HE575208) (3). With the rapid growth in bacterial genome sequencing, many new clusters are located within much larger genome sequence files and are occasionally unannotated, such as the antibiotic TA/myxovirescin biosynthetic gene cluster in the Myxococcus xanthus genome (accession number CP000113.1) (4). These problems are compounded by the fact that gene cluster discovery is being undertaken by researchers from diverse fields of expertise, including chemistry, biochemistry, microbiology, biotechnology and drug discovery, all with differing standards for gene cluster annotation. Thus, it is no surprise that given these issues, it can be extremely challenging, time consuming and often frustrating to find appropriate genes cluster in the NCBI database.
To accelerate research and leverage existing data in PKS/NRPS biosynthesis, a focused and comprehensive database that gathers this gene cluster information together is required (5). Although there are some existing databases that provide important resources on PKS/NRPS gene clusters and/or their products (6–9), none have the features necessary to enable the community to maximize the benefit from sequence data. In particular, we have identified two key features that are required for the community. The first is to have a comprehensive up-to-date database. Because of the rapid emergence of new gene clusters across a broad range of disciplines, a resource that can be easily updated by any and all community members is required to ensure that the database is comprehensive and current. The second is that the difficulty in accessing multiple diverse gene clusters has limited the ability of researchers to carry out comprehensive phylogenetic and functional analysis. Therefore, the database must have the ability to generate multiple sequence FASTA files for individual catalytic domains found in PKS and NRPS biosynthesis.
In evaluating the existing PKS/NRPS databases, we found that some of them, such as NRPS-PKS (6) and MAPSI (7), have not been updated in recent years. Others, such as NORINE (8), focus on the products of the cluster and do not contain information on the gene cluster itself. DoBISCUIT (http://www.bio.nite.go.jp/pks/) is a new and promising database, but currently has limited amounts of data, whereas PKMiner (9) is limited to type II PKS clusters. Curated databases, such as those mentioned previously, can offer high levels of data quality, but they are not always actively updated, as few institutions or research groups have the resources to maintain ongoing manual curation. Additionally, there can be long lag times between the discovery of a new gene cluster and its inclusion in a traditionally curated database. Newly discovered clusters are often excluded from these databases, as they do not meet curation criteria. For example, they may lack a characterized product as is seen for a large number of cryptic or silent gene clusters from whole genome sequencing efforts (10). The result of this is a bias towards a limited number of well-known archetypical clusters, such as the erythromycin (accession number AY623658) (11) and tyrocidine (accession number AF004835) (12) gene clusters. This is a particularly important concern for researchers attempting to assign function to new gene clusters and those involved in bioprospecting, as they need access to the breath and diversity of sequenced clusters and not simply the well-known prototypical textbook clusters. The best way to address these issues, which are limiting the research ability of the community, is to build a dynamic resource that allows users to make contributions, minimizes the amount of time-consuming manual curation by database administrators, but maintains the high standard of curated data quality.
New data, especially from bacterial genome sequencing, is being generated at an extraordinarily rapid rate (5). To keep up with this influx of data, while at the same time minimizing the amount of inefficient data entry, we chose to develop a server based workflow engine to assist in curation of gene cluster data. Additionally, we have adopted a community-based approach for the collection of data for this database. Researchers can sign up for a free account, allowing them to add to or update the database. This crowd-sourcing allows participation by those who are most interested in using the data, ensuring broad coverage of the data across diverse fields, while decreasing the need for a dedicated full-time curator.
Community-based curation has some unique challenges. In particular, it can be difficult to ensure high levels of data quality (13,14). To address this issue, we have limited the input from the users, such that only a few key details need be provided with the bulk of the data collection and analysis being performed in an automated fashion using known databases, such as the NCBI databases, and analysis tools, including antiSMASH (antibiotics and secondary metabolite analysis shell) (15). The use of automation means the database can ‘auto-curate’ itself, reducing the amount of administrative burden and enabling the database to grow dynamically through community contributions.
DATABASE ORGANIZATION
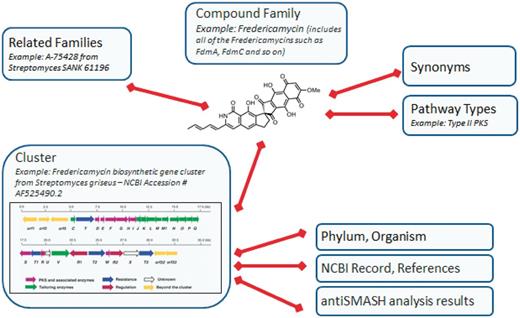
The microbial PKS/NRPS database, ClusterMine360 (http://www.clustermine360.ca/), is organized around two key elements, the compound family and the gene cluster (see Figure 1). A compound family is a grouping of compounds that have the same core structure. This term is used, as most gene clusters produce more than one compound, although they tend to be highly related. For example, the epothilone biosynthetic pathway produces four highly related polyketides, epothilones A–D, which differ by the presence or absence of a methyl group and an epoxide moiety (17,18). Thus, by organizing by compound family, we are able to capture the chemical diversity generated by a single biosynthetic gene cluster without duplicating data in the database. The ‘Compound Families’ page of the website has a listing of all of the families along with an image of the structure of a representative member of the family (if available).
Organization of ClusterMine360. The compound family and cluster represent the two major organization units of the database. Additional data fields connect to either the compound family or cluster. The organization of the fredericamycin gene cluster is shown in the cluster pane (16).
As many natural products are known by more than one name, synonyms for each compound family can be added. This is essential to limit duplicate entries. For example, the polyketide pimaricin is also widely known as natamycin. Before adding a new compound family, the list of existing names is checked to ensure it has not already been added. If the compound family has already been added under another name, the user is notified and is given the primary name for that family in the database. Additionally, the database queries ChemSpider to identify synonyms for each compound family and adds these to the compound family’s details page, ensuring a comprehensive set of synonyms for each compound family.
Because many compounds can be highly related, yet clearly not from the same compound family, each compound family can be linked to related families. For example, erythromycin, megalomycin and oleandomycin all share the same polyketide core, but differ in their sugar residues attached to the core. These are clearly highly related compounds; thus, they are linked together as related families. Identification of related families is highly subjective. Although it is possible to evaluate similarity between structures using mathematical coefficients, such as the Tanimoto similarity or Euclidian distance (19), no weighting scheme that captured the subjective relatedness of, for example, erythromycin, megalomycin and oleandomycin, without including, for example, methylmycin, narbomycin, pikromycin or lankamycin, was available. Compound families can also be related by similarities in the clusters that produce them. As part of the analysis undertaken by antiSMASH, it searches for similar clusters, and the results of these are then used to automatically link the compound families. Links to related compound families are shown on the compound family’s details page, enabling users to easily access data for related compounds. To capture some of the broader relatedness between compound families, each family is associated with one or more overall biosynthetic pathway type, such as PKS type I, type II, type III or NRPS. Clusters with PKS and NRPS domains are identified as hybrid pathways. This enables the compound families to be rapidly sorted by a broad structural relatedness.
The second major organization unit of the database is the gene cluster. Multiple clusters can be associated with a given compound family. For example, epothilone biosynthetic gene clusters have been sequenced from two strains of Sorangium cellulosum (20,21), and erythromycin gene clusters have been sequenced from Saccharopolyspora erythraea (22) and Aeromicrobium erythreum (11). Each cluster is associated with an NCBI nucleotide record. The NCBI record is used as the source for the lineage of the producing organism, including the phylum, genus and species. Links to primary literature references for the sequencing data are also retrieved from the NCBI record and displayed on the cluster’s details page. Linked to each gene cluster is the annotation data for each gene in the cluster and each domain found in the PKS and NRPS encoding genes. These data are generated through antiSMASH analysis of each gene cluster (15). The domain sequences, extracted from the antiSMASH results, are also available from the gene cluster’s details page.
AUTOMATION
Ensuring high data quality is time consuming, and it makes database upkeep difficult. One of the most important requirements for the database was to integrate automation to make curation as easy as possible. As most of the data are populated automatically, external users are able to contribute without much risk to data quality. This semi-automatic curation also means that large amounts of data can be added to the database in a relatively short amount of time.
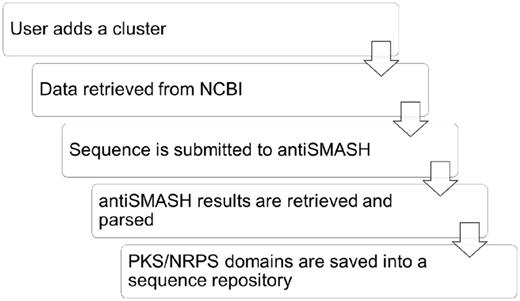
The following steps occur once a cluster is added (see Figure 2). First, the NCBI nucleotide database is queried to retrieve important information about the sequence, such as its description, the name and lineage of the organism it was isolated from and any sequencing references that are associated with the record. Once this information has been retrieved, the cluster is submitted to antiSMASH for analysis. The database automatically tracks the progress of the antiSMASH submission and proceeds to download the results when completed. The results are then parsed to retrieve information, such as the pathway types for that cluster, which is used to ensure that the pathway types of the linked compound family are correct. Finally, if antiSMASH has identified any PKS/NRPS domains, the amino acid sequence of those domains will be stored in the database’s sequence repository along with key information, such as domain substrate specificity, stereochemistry and activity of the domain, as applicable. In addition, when a compound family is added, it is searched against the PubChem (23,24) database to retrieve Medical Subject Heading (MeSH) pharmacological identifiers that classify the compound’s bioactivity. Simplified molecular-input line-entry system (SMILES) strings are also retrieved enabling users to search the database by substructure. The typical time to complete these processes ranges from a few minutes to a few hours depending on server load.
ClusterMine360 has automated many of the steps required for curating the database. Automated curation is essential to enable crowd-sourcing without sacrificing data quality.
In addition to the automated processes above, we also incorporated some other features that make it particularly easy for users to add data. When a compound family is added to the database, a wizard guides the user through the process of entering information on pathway types, synonyms and related families and helping the user in generating an image for the structure of the compound. To make it easy to associate an image, the ChemSpider database (http://www.chemspider.com) is queried to retrieve images that match the compound family name. Alternatively, an image can be generated from a user supplied SMILES string. Similarly, when adding synonyms, potential synonyms are returned from ChemSpider and the user can easily select those that are applicable.
antiSMASH
antiSMASH is the bioinformatics tool we use to provide analysis on clusters. antiSMASH can scan a cluster’s sequence and determine the most likely pathway type for that cluster. For type I PKS clusters, it also attempts to predict whether it is modular, iterative or has trans-acyltransferase (ATs). It is also able to make predictions for individual domains. It endeavours to determine the substrate specificity for AT and adenylation domains. For ketoreductase (KR) domains, it assesses whether it is active or inactive, and the probable stereochemistry of the product. More details can be found in (15). To ensure the standardization of the large amounts of data in the database and to minimize manual curation, the results retrieved from antiSMASH by ClusterMine360 cannot be edited by individual users to include new biochemistry. However, as newly characterized PKS/NRPS domains are added to antiSMASH, the clusters in the database can be easily re-analysed to take advantage of the improved analytics.
USER CONTRIBUTIONS
User contributions to the database are encouraged and acknowledged. To contribute to the database, users must register for a free account using a simple registration form. The name of the contributor, the name of their research group and a link to their webpage is displayed on records that they have added to the database.
PRESENT CONTENT
Currently, the database has >185 unique compound families, >200 clusters with known products and >300 clusters with no known products (silent or cryptic gene clusters). The sequence repository has 10 000+ PKS/NRPS domains from >500 clusters available for download, including 1300+ acyl carrier proteins (ACPs), 1000+ ATs, 1000+ KRs, 1300+ ketosynthases (KSs), 250+ thioesterases (TEs), along with sequences from less common domains, such as heterocyclization and epimerization domains.
SEQUENCE REPOSITORY
One of the most unique aspects of this database is its sequence repository. The repository contains large number of diverse PKS/NRPS domains extracted from the antiSMASH analysis of the clusters contained in the database. We have also included the ability to scan any NCBI nucleotide record and have the detected PKS/NRPS included in this repository. We believe that this repository will become an invaluable tool to those involved in identifying sequence homologies and bioprospecting. The sequences can be downloaded individually in FASTA format. Alternatively, all of the domains in a given cluster can be downloaded at once in a zip file. We have also included the ability to filter the domains based on a variety of criteria, following which they can be downloaded in a multi-sequence FASTA file. Importantly, the depth of information included in each sequence’s header is exceptional. They are full of rich information, such as accession number, producing organism, gene identifier, pathway type, domain type and any predicted properties of that domain. We have also included an option to output shortened headers for use with bioinformatics tools that have restrictions on the number of characters in the header.
ClusterMine360: A POWERFUL TOOL FOR PHYLOGENETIC ANALYSIS
To demonstrate the utility of the ClusterMine360 database, NRPS heterocyclization domains were selected and used for cluster analysis. Heterocyclization domains play a key role in NRPS biosynthesis, coupling acyl and peptidyl groups onto Cys, Ser and Thr residues followed by cyclization of the associated side-chain to generate thiazol and oxazole rings (25–27). This occurs during the biosynthesis of non-ribosomal peptides, such as the antibiotic bacitracin, and mixed non-ribosomal peptide/polyketides, such as the antimitotic agents epothilone and rhizoxin.
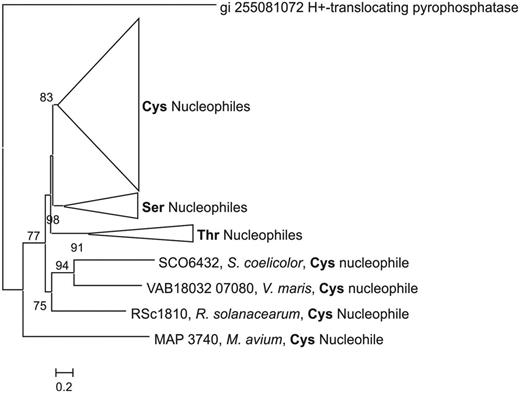
A FASTA file of 106 heterocyclization domains was downloaded and aligned using Multiple Sequence Comparison by Log-Expectation (MUSCLE) (28). A phylogenetic tree was generated from the resulting alignment using the PhyML maximum likelihood method with the Whelan and Goldman (WAG) model of amino acid substitution and nearest neighbour interchange for the tree topology search (29). The tree shows that heterocyclization domains clustered by function, based on whether the domain used enzyme bound Cys, Ser or Thr as its substrate (Figure 3). To evaluate which residues each heterocyclization domain used, the ‘detail of cluster’ function in the sequence repository was examined to identify the specificity of adenylation domain associated with the heterocyclization domain. Based on this analysis, the tree shows that Cys, Ser and Thr specific heterocyclization domains all tree apart from each other. This analysis shows that with ClusterMine360, it is possible to rapidly develop phylogenetic tools to predict the function of an individual domain.
A rooted phylogenetic tree of heterocyclization domains from NRPS gene clusters shows that heterocyclization domains tree is based on function. ClusterMine360 provides a rapid and powerful tool for generating and analysing phylogenetic trees of PKS and NRPS domains.
CONCLUSION
ClusterMine360 (http://www.clustermine360.ca/) is a unique database of microbial PKS/NRPS clusters. It contains >200 clusters from >185 compound families, and it features a unique sequence repository containing >10 000 PKS/NRPS domains. By leveraging automation and crowd-sourcing, we believe that this database will grow dynamically through contributions from interested parties as new clusters are discovered and sequenced. We are confident that this database will be a useful resource for members of the PKS/NRPS research community, enabling them to keep up with the growing number of sequenced gene clusters, and rapidly mine these clusters for functional information.
FUNDING
The National Science and Engineering Research Council of Canada (NSERC); Ontario Ministry of Research and Innovation; University of Ottawa. Funding for open access charge: University of Ottawa and NSERC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the antiSMASH development team for providing an excellent tool to the natural products community. They would like to thank Kai Blin, in particular, for his assistance in integrating the database with antiSMASH. They would also like to thank the team at GGA Software Services for creating and maintaining the Indigo open-source chemistry toolkit (http://ggasoftware.com/opensource/indigo), which is used for substructure searching and for generating images from SMILES strings. In addition, they would like to thank Dr Paul Thiessen from NIH/NLM/NCBI for providing invaluable assistance with regards to interacting with the PubChem REST interface.




Comments