-
PDF
- Split View
-
Views
-
Cite
Cite
Simonne Longerich, Youngho Kwon, Miaw-Sheue Tsai, Aye Su Hlaing, Gary M. Kupfer, Patrick Sung, Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA, Nucleic Acids Research, Volume 42, Issue 9, 14 May 2014, Pages 5657–5670, https://doi.org/10.1093/nar/gku198
Close - Share Icon Share
Abstract
FANCD2 and FANCI function together in the Fanconi anemia network of deoxyribonucleic acid (DNA) crosslink repair. These proteins form the dimeric ID2 complex that binds DNA and becomes monoubiquitinated upon exposure of cells to DNA crosslinking agents. The monoubiquitinated ID2 complex is thought to facilitate DNA repair via recruitment of specific nucleases, translesion DNA polymerases and the homologous recombination machinery. Using the ubiquitin conjugating enzyme (E2) UBE2T and ubiquitin ligase (E3) FANCL, monoubiquitination of human FANCD2 and FANCI was examined. The ID2 complex is a poor substrate for monoubiquitination, consistent with the published crystal structure showing the solvent inaccessibility of the target lysines. Importantly, FANCD2 monoubiquitination within the ID2 complex is strongly stimulated by duplex or branched DNA, but unstructured single-stranded DNA or chromatinized DNA is ineffective. Interaction of FANCL with the ID2 complex is indispensable for its E3 ligase efficacy. Interestingly, mutations in FANCI that impair its DNA binding activity compromise DNA-stimulated FANCD2 monoubiquitination. Moreover, we demonstrate that in the absence of FANCD2, DNA also stimulates FANCI monoubiquitination, but in a FANCL-independent manner. These results implicate the role of a proper DNA ligand in FANCD2 and FANCI monoubiquitination, and reveal regulatory mechanisms that are dependent on protein–protein and protein–DNA interactions.
INTRODUCTION
Interstrand deoxyribonucleic acid (DNA) crosslinks (ICLs), induced by chemicals or endogenous agents, are highly deleterious because they interfere with processes, such as DNA replication or transcription, that entail DNA strand separation. ICL removal is a major role of FANCD2 and a collection of partner proteins whose deficiency can lead to the human disease Fanconi anemia (FA), characterized by acute cellular sensitivity to DNA crosslinking agents, developmental abnormalities, cancer susceptibility and bone marrow failure (1,2). Upon treatment of cells with a DNA crosslinking agent, e.g. mitomycin C and cisplatin, FANCD2 is monoubiquitinated on lysine 561 and it becomes associated with damaged DNA. Monoubiquitinated FANCD2 co-localizes with a number of proteins that possess nuclease activity or that promote DNA repair by homologous recombination or translesion DNA synthesis (3–5). Several such proteins with ubiquitin binding motifs are recruited to damaged DNA via their interaction with monoubiquitinated FANCD2 (6–9).
FANCD2 associates with FANCI to form the heterodimeric ID2 complex (10,11). FANCI is structurally related to FANCD2 (10) and, like FANCD2, is monoubiquitinated (on lysine 523) in response to DNA damage. Monoubiquitinated FANCI localizes to DNA damage foci and is required for the cellular resistance to DNA crosslinking agents (12–14).
Interestingly, the recently published crystal structure of the murine ID2 complex shows that the lysine residues targeted for monoubiquitination in FANCD2 and FANCI are buried in the dimer interface and are expected to be unavailable for modification (10). An important question is thus how the monoubiquitination sites in the ID2 complex are rendered accessible to the ubiquitin conjugation machinery during the activation of the FA pathway of DNA damage response and repair.
Notably, both FANCD2 and FANCI possess DNA binding activity, and the latter exhibits preferential binding to branched DNA substrates that resemble DNA repair intermediates (10,15,16). How a DNA ligand may influence the monoubiquitination of FANCD2 and FANCI is of great interest because of the essential function these modified proteins perform in ICL repair. To address this question, we have developed a reconstituted system consisting of only highly purified protein components to examine the monoubiquitination of human ID2 complex by its cognate E2 ubiquitin conjugating enzyme UBE2T and E3 ligase FANCL. We show that ubiquitination of FANCD2 and FANCI within the context of the ID2 complex is minimal, but the addition of an appropriate DNA substrate greatly stimulates FANCD2 monoubiquitination and may also enhance FANCI monoubiquitination. Mutations in FANCI that compromises its DNA binding attribute attenuate the stimulatory effect of DNA on ID2 ubiquitination, as do two patient-derived mutations that are located at the C-terminus of FANCI. Furthermore, despite a direct interaction of FANCD2 with free histones, ID2 ubiquitination is suppressed when the DNA is nucleosome bound. Finally, even though the monoubiquitination site in FANCI is apparently solvent exposed in the absence of FANCD2 (10), we demonstrate that monoubiquitination of FANCI is also stimulated upon DNA binding. Altogether, the results reveal that an appropriate DNA ligand induces a conformational change in FANCI and the ID2 complex that is conducive for their monoubiquitination. Moreover, the results identify FANCI as a sensor in the signal relay from DNA binding to protein monoubiquitination.
MATERIALS AND METHODS
Cloning
The primers used in complementary DNA (cDNA) subcloning and mutant generation are listed in Supplementary Table S2. FANCL cDNA was amplified by polymerase chain reaction (PCR) from pDEST20-FANCL (15) with primers FL1 and FL2 and was inserted between the EcoRI and SalI sites in pMAL-TEV (17) to yield pMAL-TEV–FANCL. The FANCL W212A/L214A mutant was constructed by replacing the ClaI–SalI fragment of pMAL-TEV–FANCL with an FANCL PCR product (using FL3 and FL2 primers) that harbors the double mutation. pFastBac-FANCI, described previously (15), was modified by QuikChange mutagenesis to generate the R1285Q, R1285X, K294E and K339E mutations. An N-terminal 3X-Flag tag was introduced into FANCD2 in the vector pAS2.1 (18). The FANCD2 K561R mutation was produced by QuikChange mutagenesis with primers FD1 and FD2. The tagged FANCD2 sequences were removed from the resulting plasmid by restriction digest and were inserted into pFastBac-Dual (Invitrogen). Bacmids harboring wild-type and mutant FANCD2 and FANCI were produced in Escherichia coli DH10Bac cells according to the manufacturer's instructions (Invitrogen). Sf9 insect cells (Invitrogen) were transfected with bacmids to produce baculoviruses and were also used to amplify the viruses.
Protein expression and purification
MBP-FANCL and the MBP-FANCL W212A/L214A mutant were produced in E. coli Rosetta DE3 pLysS (Novagen) in 2XLB medium containing 0.5-mM ZnCl2. When cells reached OD600 of 0.8–1, protein expression was induced with 0.1-mM isopropyl-beta-D-thiogalactopyranoside (IPTG) for 16 h at 16°C. Cell pellets were frozen at −80°C. FANCI, the ID2 complex and their mutant variants were produced in High Five insect cells by infecting cells with our baculoviruses and harvesting the cells 48–50 h post infection. Insect and E. coli cell pellets were lysed by sonication and cleared by high-speed centrifugation (100 000 × g, 60 min).
For the purification of MBP-FANCL and the MBP-FANCL W212A/L214A mutant, cells were lysed in buffer A (25-mM Tris, pH 7.5, 10% glycerol, 1-mM ethylenediaminetetraacetic acid (EDTA), 1-mM β-mercaptoethanol and 0.01% NP-40) supplemented with 300-mM KCl and 0.5-mM ZnCl2, and the lysate was clarified by ultracentrifugation. The lysate was then subjected to affinity chromatography using Amylose resin (New England Biolabs) followed by size exclusion chromatography using a Superdex 200 column (GE Healthcare) in the same buffer. Fractions containing unaggregated MBP-FANCL were pooled, concentrated in an Amicon Ultra-4 centrifugal filter device, and frozen in small aliquots at −80°C.
FANCI and its mutant variants were purified from insect cells as described (15) with the addition of a terminal step of size exclusion on a Superdex 200 column using buffer A with 300-mM KCl.
The ID2 complex was purified from insect cells co-infected with baculoviruses that produce either wild type or mutant FANCI and FANCD2. Cells were lysed in buffer A containing 120-mM KCl and supplemented with RNaseA (2 μg/ml), cleared and purified using anti-Flag (Sigma) and Nickel-NTA (Qiagen) affinity steps, followed by size exclusion chromatography on a Superdex 200 column in buffer B (25-mM Tris, pH 7.5, 75-mM KCl, 2% glycerol and 1-mM EDTA supplemented with 1-mM β-mercaptoethanol and 0.01% NP-40). Fractions containing the ID2 complex were pooled, concentrated and stored in small aliquots at −80°C.
hUBE1 and haemagglutinin (HA)-ubiquitin were purchased from Boston Biochem. The expression and purification of UBE2T and the UBE2T C86A mutant were exactly as described previously (15).
DNA substrates
ϕX174 RFI, RFII and virion were purchased from New England Biolabs. Oligonucleotides used in the monoubiquitination assays were purchased from IDT, purified (15) and stored in TE buffer (10-mM Tris-HCl, pH 8.0 and 1-mM EDTA). DNA substrates were prepared by mixing the relevant oligonucleotides (Supplementary Tables S1 and S3) in equimolar amounts in buffer C (50-mM Tris-HCl, pH 7.5, 10-mM MgCl2, and 100-mM NaCl), boiling for 10 min and then cooling gradually to room temperature. The substrates were precipitated with ethanol and suspended to a final concentration of 25 μM in TE buffer.
Chromatinized DNA substrates
Plasmid DNA (pBluescript SK) was assembled into polynucleosomes with core histones as described (19) or treated without histones to serve as the control substrate. Briefly, chicken histone octamers were mixed with the DNA in TE buffer containing 1.5-M NaCl, incubated on ice for 45 min, whereupon the mixture was dialyzed stepwise against TE buffer containing progressively lower concentrations of NaCl until reaching a final concentration of 25-mM NaCl. Nucleosome core particles were prepared as described (20).
Ubiquitination assays
The monoubiquitination assays were performed as described (15) but using MBP-FANCL instead of GST-FANCL and with the addition of DNA. MBP-FANCL has up to ∼3-fold higher specific activity than the previously described GST-FANCL. Briefly, FANCI or the ID2 complex (0.2–0.4 μM) was mixed with DNA in the reaction buffer (50-mM Tris-HCl, pH 7.5, 22.5-mM KCl, 4-mM MgCl2, 0.01-mg/ml bovine serum albumin and 0.5-mM (Dithiothreitol)DTT) and incubated at room temperature for 10 min. The reaction tubes were returned to ice for 2 min whereupon the following components were added: 0.1-μM UBE1, 0.3–0.7-μM UBE2T, 0.3-μM MBP-FANCL, 32-μM HA-ubiquitin and 2-mM adenosine triphosphate. The salt concentration in the reactions was 67 mM in a final volume of 12.5 μl. The reaction mixtures were incubated at room temperature for the indicated times and terminated by the addition of 5 μl of buffer [6% sodium dodecyl sulphate (SDS), 0.25-M Tris-HCl, pH 6.8, 50% glycerol, 2.7-M β-mercaptoethanol, with bromophenol blue], followed by electrophoresis in 6.5% SDS polyacrylamide gels. The gels were stained with Oriole Fluorescent Gel stain (BioRad) and analyzed in a gel documentation system (BioRad) fitted with an ultraviolet source. Ubiquitinated products were quantified using ImageQuant software on images from the Oriole-stained gels. The reaction mixtures (10–15% total) were also analyzed by western blotting with horseradish peroxidase-conjugated anti-HA (Sigma).
DNA binding assays
DNA mobility shift reactions were assembled in the buffer used in the ubiquitination assay and analyzed according to our published protocol (15).
RESULTS
Preparation of FANCI and the ID2 complex for monoubiquitination studies
We reported previously the expression of FANCI with the use of a recombinant baculovirus in insect cells and also a method for its purification to near homogeneity (15). FANCI thus expressed and purified possesses a robust DNA binding activity and can be monoubiquitinated on the physiological site (i.e. K523), albeit only poorly (15). As we will document below, the monoubiquitination of FANCI, either alone or in complex with FANCD2, is stimulated by certain DNA types.
We employed a similar strategy to express FANCD2 in insect cells. Even though FANCD2 thus expressed is soluble, it is highly prone to aggregation. Many batches of FANCD2 were prepared, but all of them, either alone or when mixed with purified FANCI, were refractory to ubiquitination with and without DNA present.
To prepare the ID2 complex, insect cells were co-infected with the FANCI and FANCD2 baculoviruses, which produces a soluble ID2 complex that could be purified to near homogeneity with a good yield. The purified ID2 complex harbors stoichiometric amounts of FANCD2 and FANCI and exhibits a mean size of ∼400 kDa in gel filtration analysis, and is thus heterodimeric. As we will document below, the monoubiquitination of the complex is enhanced by certain DNAs.
Enhancement of ID2 monoubiquitination by DNA
All the ubiquitination assays described herein were conducted using maltose-binding protein (MBP)-tagged FANCL, which is more amenable to purification than the previously described glutathione-S-transferase (GST)-tagged protein (15) and has a significantly higher specific activity than the latter (data not shown).
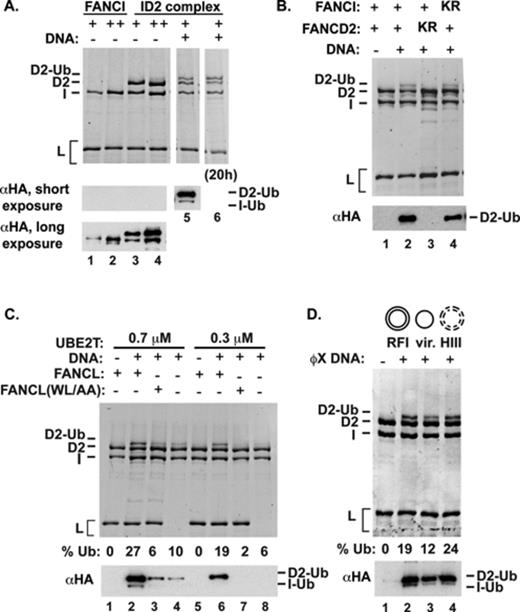
We conducted many experiments using several different preparations of the purified ID2 complex and found it to be a poor substrate for UBE2T-FANCL. We note that this result is consistent with the crystal structure of the murine ID2 complex in which the target lysine residues in FANCD2 and FANCI are buried within the interface between the two proteins. Since both FANCD2 and FANCI possess a DNA binding activity (10,11,15,16,21), we then investigated how DNA might affect their ubiquitination. Interestingly, addition of plasmid DNA greatly stimulated the formation of ID2 ubiquitination products, the major form of which (later identified to be monoubiquitinated FANCD2) could be detected easily by staining with a fluorescent dye after sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 1A, lanes 5 and 6, and Supplementary Figure S1). The analyses presented in Supplementary Figure S1 also revealed that the various protein components remained stable over the course of the ubiquitination reaction. Since the ubiquitin used in our reconstituted system is tagged with the HA epitope, we could also visualize the ubiquitinated ID2 species by immunoblot analysis using a commercially available anti-HA antibody. The immunoblot analysis revealed, aside from the predominant species, a minor one that possesses a faster mobility. The identity of the two ubiquitinated species was established by substituting the ID2 complex with variants that harbor either the FANCD2 K561R or FANCI K523R mutation. As summarized in Figure 1B (and see later), the analysis provided direct evidence that (i) DNA-stimulated ubiquitination occurs site specifically on FANCD2 K561 and FANCI K523, (ii) FANCD2 ubiquitination is mostly or completely independent of FANCI ubiquitination and (iii) with DNA present, ubiquitination of FANCI is less robust than FANCD2 modification in the ID2 complex. We focus first on the mechanism of FANCD2 ubiquitination in the ID2 complex, followed by further dissection of FANCI ubiquitination below.
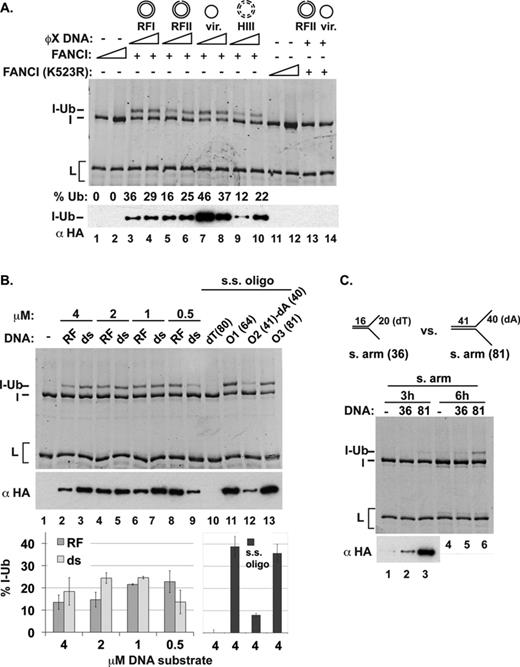
Enhancement of ID2 monoubiquitination by DNA. (A) FANCI or the ID2 complex was used as substrate in ubiquitination reactions. ϕX RFI DNA (45 nM) was added to the reaction with the ID2 complex in lanes 5 (3-h reaction) and 6 (18-h reaction). Fluorescent staining of proteins is shown on the top, and two different exposures of a western blot with α-HA are shown underneath. (B) ID2 complexes containing wild-type proteins, FANCD2 (K561R) or FANCI (K523R) were tested. (C) ID2 ubiquitination was examined in the presence of wild-type or mutant FANCL or without FANCL. (D) ID2 ubiquitination was tested with various forms of ϕX DNA (RF-replicative form; HIII-HaeIII digested RF DNA). For (B)–(D), fluorescent-stained images on top are from 18-h reactions, whereas the western blots show products from 3-h reactions.
Maximal FANCD2 ubiquitination shows a strong dependence on FANCL in our reconstituted system (Figure 1C). FANCL interacts transiently with FANCD2 and FANCI, and a hydrophobic patch in the middle portion of FANCL has been shown to be important for these protein interactions (22). Mutation of the W212 and L214 residues located in this hydrophobic patch attenuates the ability of FANCL to interact with FANCD2 and FANCI (22). We have constructed, expressed and purified the FANCL W212A, L214A mutant protein and tested it in the ID2 ubiquitination reaction. The results revealed that interaction with the ID2 complex is critically important for FANCD2 ubiquitination (Figure 1C).
We initially used different forms of ϕX174 plasmid DNA to determine whether supercoiling (compare RFI versus RFII) or single-stranded nature (compare the viral form with other forms) or the number of available double-stranded DNA ends (compare HaeIII-digested DNA with other forms) would differentially stimulate ID2 ubiquitination, but no significant difference among the various substrates was detected (Figure 1D).
Aside from FANCD2 and FANCI, the other proteins in the ubiquitination reaction do not bind DNA. These results therefore revealed that when associated with DNA, the ID2 complex adopts a new conformation in which K561 of FANCD2 becomes accessible for ubiquitination.
Chromatin is repressive for DNA-stimulated ID2 ubiquitination
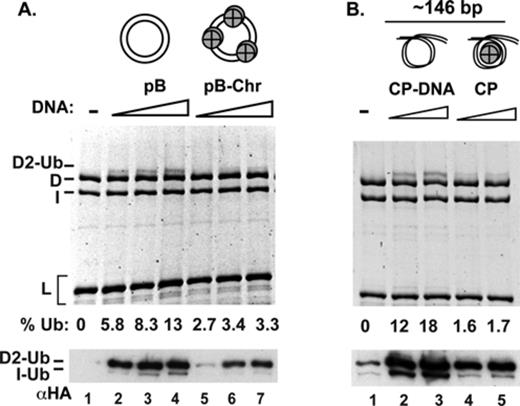
Chicken FANCD2 protein binds the histone H3/H4 complex (23). Moreover, the ubiquitinated form of FANCD2 and FANCI is enriched in the chromatin fraction of cells (3,12,13). To ask whether chromatin is a more proficient co-factor of ID2 monoubiquitination, we tested the efficacy of a nucleosomal array (Supplementary Figure S2A) in the ubiquitination reaction. Even though the nucleosomal DNA template was able to stimulate FANCD2 ubiquitination, the nucleosome-free DNA was more effective, by a factor of two to three (Figure 2A).
Comparison of nucleosomal DNA and free DNA in ID2 ubiquitination. (A) Nucleosomes assembled on plasmid DNA, or (B) in the context of core particles, were compared to free DNA in ID2 ubiquitination. The reaction mixtures were analyzed by fluorescent staining of proteins after 6 h of incubation (top) or by western blotting after 1.5 h of incubation (bottom). The percent of FANCD2 that had been ubiquitinated (% Ub) after 6 h of incubation is indicated.
To further test the idea that free DNA, not chromatinized DNA, is the physiologically relevant co-factor in ID2 ubiquitination, we compared nucleosome core particles (mononucleosomes, each wrapped around ∼150 base pairs of DNA) against the same DNA that had been released from the particle in the monoubiquitination reaction (Supplementary Figure S2B). Again, nucleosome-free DNA stimulated FANCD2 ubiquitination more efficiently than the core particle, by a factor of 8 to 10 (Figure 2B). By DNA mobility shift, we additionally found that the ID2 complex binds less well to the core particle versus the naked DNA released from the core particle (Supplementary Figure S2C).
DNA substrate specificity in ID2 ubiquitination
Results presented above (Figure 1D) revealed that plasmid length double- and single-stranded DNAs are equally effective in the promotion of ID2 ubiquitination. However, these DNAs are long, with undefined secondary structures. To address how specific DNA sequence context and structures influence the efficacy of the DNA ligand in the ubiquitination reaction, we determined the relative affinity of the purified ID2 complex for various oligonucleotide-based substrates, including single- and double-stranded DNAs as well as branched substrates that mimic a stalled replication fork, and also the relative efficacy of these DNA substrates in the reconstituted ubiquitination reaction.
In cell-free DNA crosslink repair reactions, FANCD2 helps promote DNA cleavage and removal of ICLs upon collision of the replication machinery with the lesion, i.e. a stalled fork (24) which represents a potential specific DNA substrate for the ID2 complex. However, unlike FANCI (15,16), the purified ID2 complex does not appear to bind fork substrates preferentially over linear double-stranded DNA (16) (Supplementary Figure S3A). Interestingly however, single-stranded oligonucleotides with predicted secondary structure are preferentially bound over an 80-nucleotide dT oligomer (Supplementary Figure S3B and C).
All the substrates that contained at least 41 base pairs of double-stranded DNA stimulated FANCD2 ubiquitination with a comparable efficiency (Figure 3A). Notably, the same splayed-arm substrate (20 base pairs, with 2 oligo-dT arms of 16 nucleotides each), used to form a co-crystal with FANCI (10), poorly stimulated FANCD2 ubiquitination (Supplementary Figure S4). In summary, substrates with at least 41 base pairs of linear double-stranded DNA stimulate FANCD2 ubiquitination, but those with 20 base pairs of double-stranded DNA are ineffective.
DNA substrate specificity in ID2 ubiquitination. (A) ID2 ubiquitination was examined with a variety of oligonucleotide-based DNA substrates as indicated (either 0.4 or 4 μM) or with 4-μM single-stranded oligonucleotides [lanes 12–13 and (B)]. Analysis was by either fluorescent staining (18-h incubation) or western blotting (3-h incubation). Quantification of FANCD2 monoubiquitination after 18 h of incubation is shown. Error bars indicate the standard deviation from at least two independent experiments.
Initially, we tested two single-stranded oligonucleotides (O2 and O3) that were used in the construction of the various substrates; one of these stimulated FANCD2 ubiquitination very well (O3) and the other only poorly (O2). These results prompted us to examine a series of single-stranded oligonucleotides that differ in sequence and length (Figure 3B). We found that the 80-nucleotide dT oligomer is ineffective in supporting FANCD2 ubiquitination, while several oligomers of random sequence are fully functional in this regard. Importantly, the efficacy of the single-stranded DNA species in the FANCD2 ubiquitination reaction correlates with the predicted tendency of the oligonucleotides to form secondary structure (Supplementary Table S1, ΔG) and how well they are bound by the ID2 complex (Supplementary Figure S3B and C). In sum, we have found that FANCD2 ubiquitination is stimulated by double-stranded DNA, or by single-stranded DNA with secondary structure.
Mutations in FANCI that impair DNA binding by the ID2 complex and ubiquitination
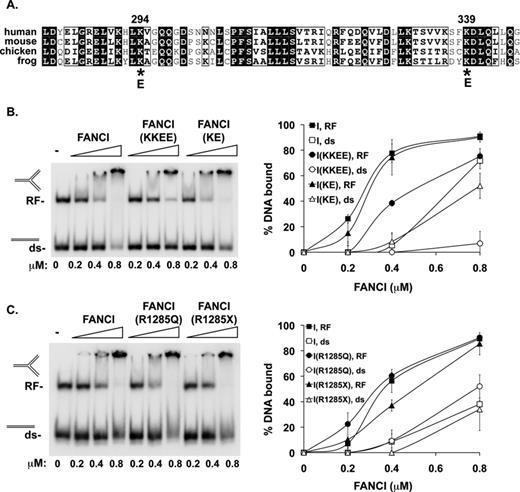
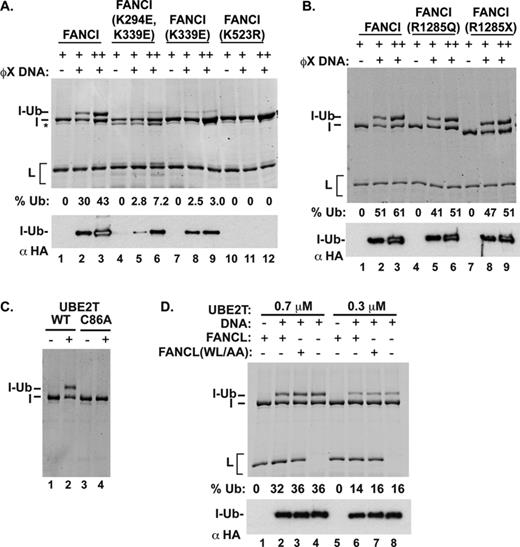
The above results revealed a dramatic stimulatory effect of certain types of DNAs on FANCD2 ubiquitination. We previously showed that FANCI possesses a robust DNA binding activity and we wished to ascertain the relevance of this activity in FANCD2 ubiquitination. To do so, we sought mutations that would compromise the DNA binding activity of the ID2 complex. Initially, we used an in silico approach based on the FANCI amino acid sequence (using PHYRE: Protein Homology/analogY Recognition Engine) to identify positively charged residues that could constitute part of the DNA binding domain. We note that the crystal structure of FANCI reveals several N-terminal, basic amino acids involved in DNA binding (10), and two of these residues, K294 and K339, were also identified in our in silico analysis. We thus elected to change these residues (Figure 4A) to glutamic acid to generate the K339E and K294E/K339E mutants. The two mutant FANCI proteins were expressed and purified like the wild-type counterpart and with a similar yield. Using the DNA mobility shift assay, we found that the K339E mutant is not deficient in DNA binding, while the K294E/K339E mutant is much more impaired in this regard (Figure 4B). We also constructed, expressed and purified two additional FANCI variants that harbor patient-derived mutations, namely, R1285Q and R1285X (a nonsense mutation at codon 1285 truncating the last 44 residues) (12,25), but discovered that only the R1285X has a slight effect on DNA binding (11,16) (Figure 4C).
Effects of FANCI mutations on DNA binding. (A) The region encompassing residues 281–346 of human FANCI is aligned against the equivalent region in several orthologs. The two lysine residues at positions 294 and 339 in the human protein selected as mutagenesis targets are highlighted. (B) The DNA binding activity of FANCI, two mutant variants (K339E and K294E/K339E, designated as KE and KKEE, respectively) and (C) two patient-derived mutant proteins (FANCI R1285Q and R1285X) were tested with 0.15 pmol each of dsDNA (81 base pairs) and a branched structure that mimics a stalled replication fork (RF; 41 base pairs of double-stranded DNA plus two double-stranded arms each of 40 base pairs). Quantification of the results is shown on the right. Error bars indicate the standard deviation from at least two independent experiments.
We then analyzed, within the context of the ID2 complex, the effects of three of these FANCI mutations (namely, K294E/K339E, R1285Q and R1285X) on DNA binding activity and ubiquitination. These FANCI mutations have no impact on complex formation with FANCD2, and we were able to assemble the respective mutant ID2 complexes by the same method as the wild-type counterpart. The analysis revealed that the FANCI K294E/K339E mutation imparts a severe DNA binding deficiency to the ID2 complex. Interestingly, the R1285Q and R1285X mutations also attenuate ID2 DNA binding (Figure 5A). Importantly, we found that all three FANCI mutations strongly impair DNA-stimulated FANCD2 ubiquitination (Figure 5B). Overall, despite the fact that FANCD2 also binds DNA (21), our results indicate that FANCI is critically important for the DNA binding affinity of the ID2 complex and, in turn, exerts a major influence on the ubiquitination of FANCD2.
Effects of FANCI mutations on ID2 ubiquitination. (A) DNA binding activity of ID2 complex and mutant variants (harboring the FANCI K294E/K339E, R1285Q or R1285X mutation) was examined as in Figure 4B. Quantification of the data is shown on the right. Error bars indicate the standard deviation of at least two independent experiments. (B) Ubiquitination reactions were carried out with the ID2 complex and its mutant variants with or without DNA [4 μM dsDNA (ds), replication fork DNA (RF) or oligonucleotide O10 (ss)] and analyzed as previously. The incubation time was 18 h for the upper panels and 3 h for the western blots.
Evidence for the enhancement of FANCI monoubiquitination by DNA
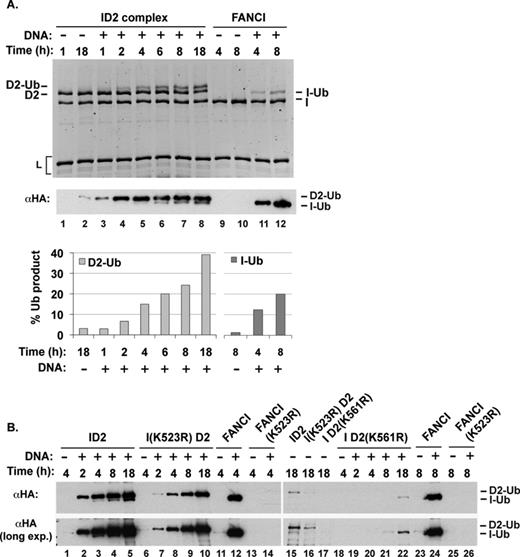
The above results revealed that within the ID2 complex, DNA induces robust monoubiquitination of FANCD2 on its cognate acceptor site (lysine 561). Curiously however, the ubiquitination of FANCI remains inefficient and delayed when compared to FANCD2 modification, as clearly visualized in a time course experiment (Figure 6). Since in cells, both FANCD2 and FANCI are ubiquitinated in response to DNA damage (12–14), we examined whether the monoubiquitination of purified FANCI alone is also stimulated by the presence of DNA. Importantly, the addition of various forms of DNA greatly enhanced FANCI monoubiquitination in the absence of FANCD2 (Figure 6A, lanes 11 and 12, and Figure 7). However, in 4 h, ubiquitinated FANCI is barely detectable in the ID2 complex, whereas in the absence of FANCD2, ∼15% of FANCI is ubiquitinated by this time (Figure 6A; compare lanes 5 and 11). Importantly, the use of the FANCI K523R mutant protein, which is proficient in DNA binding (Supplementary Figure S5), confirmed that ubiquitin is conjugated to the physiological site in FANCI with and without FANCD2 present (Figure 6B and Figure 7, lanes 11–14). These results indicate that in the ID2 complex, FANCI ubiquitination is attenuated relative to free FANCI.
(A) Time course of ubiquitination of either FANCI or the ID2 complex. Quantification of the results is shown. (B) Time course of ubiquitination of the ID2 complex, FANCI and the indicated mutant ID2 complex or mutant FANCI.
Enhancement of FANCI ubiquitination by DNA. (A) Various forms of ϕX plasmid DNA (11 or 45 nM; HIII-HaeIII digested) were added to ubiquitination reactions that contain FANCI (I) or the FANCI K523R mutant. The position of monoubiquitinated FANCI (I-Ub) is marked. The percent of ubiquitinated FANCI product (% Ub) is indicated. (B) FANCI ubiquitination reactions were carried out with linear dsDNA (ds), replication fork (RF) DNA or ss oligonucleotides (O1, O2 and O3) that differ in sequence and length. Reaction products were analyzed by SDS-PAGE and fluorescent staining (18-h incubation) or western blot (3-h incubation). Reaction product formed after 18 h of incubation was quantified. (C) FANCI ubiquitination reactions were performed as in panel (A) but comparing two splayed-arm DNA substrates that differ in length.
With plasmid-length DNA, the ϕX174 viral form appeared to be slightly more effective than the other forms in stimulating FANCI ubiquitination (Figure 7A). Since FANCI preferentially binds branched DNA substrates and single-stranded DNA with secondary structure (15,16) (Supplementary Figure S3A and Figure 4), we next compared various defined oligonucleotide-based DNA substrates to determine the DNA substrate specificity in FANCI ubiquitination. The results revealed that a linear duplex is just as effective as model replication fork structures (5’ flap, 3’ flap or RF) in supporting FANCI ubiquitination (Figure 7B). However, as with the ID2 complex, a striking difference was observed in the ability of various single-stranded oligonucleotides to promote FANCI ubiquitination. Specifically, whereas an 80-nucleotide oligomer of dT lacks activity (Figure 7B, lane 10), two random oligomers of 64 or 81 nucleotides are highly effective (Figure 7B, lanes 11 and 13), more effective in fact than double-stranded DNA (Figure 7). Interestingly, an 81-nucleotide oligomer comprised of 41 random residues adjoining 40-dA nucleotides also exhibits activity, albeit at an intermediate level (Figure 7B, lane 12). We note that, as we have observed for the ID2 complex, the ability of a particular type of single-stranded DNA to promote FANCI ubiquitination correlates very well with its propensity to form secondary structure (Supplementary Table S1, ΔG). The results suggest that engagement of a proper DNA ligand is conducive for FANCI ubiquitination. Finally, the splay-armed DNA with only 20 base pairs of double-stranded DNA used in forming a co-crystal with the murine FANCI protein (10) proved to be poorly adept at promoting FANCI ubiquitination (Figure 7C).
FANCI ubiquitination is reliant on DNA binding but not on FANCL
To ascertain the role of DNA binding in the DNA-stimulated ubiquitination of FANCI, we examined the various FANCI variants that we generated. The results showed that FANCI K339E and K294E/K339E mutant proteins are both impaired for ubiquitination (Figure 8A). In contrast, the two FA patient-derived FANCI mutations, R1285Q and R1285X (12,25), have little or no impact on the ubiquitination efficiency (Figure 8B). As expected, FANCI ubiquitination did not occur when the UBE2T active site C86A mutant was used (Figure 8C). Interestingly, however, we found that the DNA-stimulated ubiquitination of FANCI occurs just as efficiently with the FANCL (W212A, L214A) mutant (22) or upon omission of FANCL (Figure 8D, lanes 3 and 7 and lanes 4 and 8, respectively).
(A) Robust FANCI ubiquitination requires its DNA binding activity but not FANCL. Reactions with FANCI or one of its mutants (0.5 or 1 μM), and with or without DNA (45-nM ϕX virion), were incubated for 18 h (fluorescent-stained gel) or 3 h (western blot). The asterisk indicates a degradation band of FANCI. (B) The FANCI R1285X and R1285Q mutants are ubiquitinated well. The experiments were performed under the same conditions as in panel (A). (C) Ubiquitination reactions were performed with and without UBE2T or the C86A catalytically defective mutant. Analysis was as described for panel (A). (D) Ubiquitination reactions were performed with or without ϕX replicative form DNA (34 nM), FANCL or the W212A/L214A (WL/AA) mutant variant, as indicated. Analysis was as described for panel (A).
Ubiquitinated FANCI binds DNA as well as non-ubiquitinated FANCI
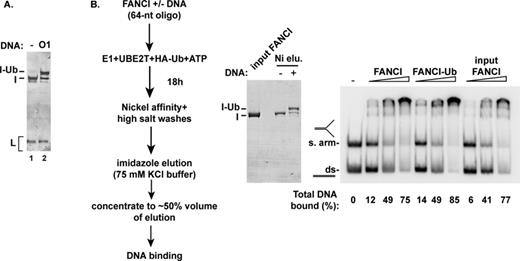
Although FANCI ubiquitination is likely required to directly recruit proteins with ubiquitin-binding domains to damaged DNA, we considered the additional possibility that ubiquitination of FANCI regulates its DNA binding activity. Using our in vitro ubiquitination system, we obtained FANCI protein that was >50% in the monoubiquitinated form. We tested this sample for DNA binding, as compared with non-ubiquitinated FANCI (either untreated or subjected to a mock ubiquitination reaction), on a mixture of two substrates: double-stranded and splayed-arm DNAs. No difference in DNA binding or substrate preference of ubiquitinated versus non-ubiquitinated FANCI was observed (Figure 9). Thus, ubiquitination is unlikely to be a mechanism for releasing FANCI from DNA.
Monoubiquitination of FANCI has no impact on DNA binding. (A) FANCI monoubiquitination (20-h incubation) was conducted with oligonucleotide O1 (64 nucleotides) as DNA co-factor. A control reaction without DNA (lane 1) was performed. (B) After affinity purification (left and middle panels), the unmodified FANCI and the monoubiquitinated preparation were tested for DNA binding using linear dsDNA and splayed-arm DNA (right panel), as in Figure 4.
DISCUSSION
Using a newly developed reconstituted system consisting of highly purified human proteins, we have provided insights into the role of DNA in the ubiquitination of the ID2 complex. Notably, we have shown that the addition of one of a variety of DNA ligands induces robust FANCD2 ubiquitination, in a manner that requires the interaction of the ID2 complex with FANCL. The monoubiquitination of FANCI is likewise stimulated by the same DNA ligands, although in the ID2 complex, the kinetics and efficiency of this reaction trail behind those that characterize FANCD2 modification. At this time, we cannot distinguish whether the lag represents a dependence of FANCI ubiquitination on prior FANCD2 ubiquitination or that the ID2 complex dissociates to release free FANCI at the later incubation time points. Nevertheless, we note that our in vitro results mirror those observed in Xenopus laevis cell-free systems in which FANCI monoubiquitination peaks at a much later time than FANCD2 modification (24,26). Moreover, we observed that FANCI monoubiquitination may be compromised by the FANCD2 K561R mutation, whereas the FANCI K523R mutation has little or no effect on FANCD2 modification. Consistent with this, FANCI ubiquitination in vivo requires FANCD2 ubiquitination, but not vice versa (12,27).
A reconstituted system for ID2 ubiquitination was recently developed using purified chicken proteins (16). In this system, the monoubiquitination of FANCD2 occurs at the physiological site and is stimulated by a variety of single-, double- and branched DNAs tested. Monoubiquitination of chicken FANCI was also observed at a low level, but it does not appear to occur on the physiological lysine (16). We note that in cell-free extracts made from X. laevis eggs, the monoubiquitination of FANCD2 at its physiological site is similarly enhanced by the addition of DNA (28). Taken together, the results from our current study and published work indicate that efficient monoubiquitination of FANCD2 and FANCI is dependent on a DNA ligand, and they support the premise that DNA binding induces a conformational change in the ID2 complex that renders the ubiquitin-targeted lysine in FANCD2, and perhaps also the target lysine in FANCI, accessible for modification (Supplementary Figure S6). Conceivably, in the context of the ID2 complex, robust levels of FANCI monoubiquitination may require other components of the FA core complex and/or phosphorylation of FANCI and FANCD2 shown to be important for FANCD2 and FANCI ubiquitination in vivo (27,29,30).
Our detailed examination of the DNA ligand specificity in ID2 ubiquitination has revealed several surprises. Most notably, a variety of DNA substrates that do not bear the features of a DNA replication fork are nonetheless highly efficient in stimulating ubiquitination. However, the use of oligonucleotide-based substrates led to the realization that the efficacy of a DNA species in the ID2 ubiquitination reaction is directly linked to the presence of secondary structure or duplex character in the DNA. Moreover, even though FANCD2 interacts with histones H3/H4, DNA packaged into nucleosomes is much less effective than naked DNA in supporting ID ubiquitination. These data support the idea that in cells, the ID2 complex need only associate with a region adjacent to a stalled replication fork or an ICL to become monoubiquitinated. The FA core complex and UBE2T reportedly are recruited to chromatin independently of one another and FANCD2 (31). Potentially, site specificity could be promoted by the coincident, proximal recruitment to damaged DNA of the ubiquitination machinery and the ID2 complex.
By testing point mutants of FANCI, we have furnished evidence that the DNA binding activity of FANCI plays a key role in regulating not only FANCD2 ubiquitination but also its own modification. Surprisingly, the two patient-derived FANCI mutations, R1285Q and R1285X, while having little-to-no effect on DNA binding by FANCI alone, exert a significant negative impact on the DNA binding affinity of the ID2 complex and, accordingly, impair ID2 ubiquitination severely. The mechanism by which these FANCI C-terminal mutations affect DNA binding by the ID2 complex remains to be determined; residues in the FANCI C-terminus, while sufficient for DNA binding of FANCI (15), appear to be necessary for DNA binding mainly in the context of the ID2 complex. The results altogether strongly suggest that FANCI acts as a crucial sensor, which adopts an altered conformation upon DNA binding that is relayed through interaction with FANCD2 to license ID2 ubiquitination.
We note that even though the target lysine in the FANCI monomer structure is fully solvent exposed as revealed by X-ray crystallography (10), its monoubiquitination remains responsive to the addition of DNA. Interestingly, the DNA-stimulated, site-specific monoubiquitination of FANCI is UBE2T dependent, but does not require FANCL. These results indicate that a DNA-induced conformational change in the FANCI monomer exposes and/or positions a UBE2T-interaction surface that promotes targeted ubiquitination of FANCI K523. A similar explanation was proposed for the DNA-induced SUMOylation of proliferating cell nuclear antigen by the E2 conjugating enzyme Ubc9 (32). We note that no obvious conformational change in FANCI was observed in the co-crystal structure of FANCI bound to a splayed-arm structure (10). However, in our reconstituted system, this same DNA is inefficient for activating FANCI ubiquitination and, therefore, may not induce the protein conformational change that leads to robust FANCI ubiquitination. FANCI therefore may be similar to a previously described class of proteins that are substrates for E3-independent ubiquitin ligation [e.g. (33)]. Since only a fraction of FANCD2 and FANCI are found in complex with each other in cells (12), these proteins may perform additional biological functions independent of their interaction in the ID2 complex. We have shown that monoubiquitination of FANCI has little or no impact on its DNA binding activity, thus ruling out this modification as a mechanism for releasing FANCI from DNA. The biological relevance of FANCL-independent ubiquitination of FANCI is an interesting question for future studies.
In summary, we have devised a reconstituted system of human ID2 ubiquitination that bears characteristics parallel to those observed for the in vivo reaction, This system should be useful for addressing mechanistic questions concerning ID2 monoubiquitination, e.g. for ascertaining the functional significance of DNA binding by FANCD2 (21) and of ATR-mediated phosphorylation of FANCI and FANCD2 (27,29,30). Moreover, our system should make it possible to obtain a sufficient amount of ubiquitinated ID2 complex for testing its role in the recruitment of repair nucleases, translesion polymerases and homologous recombination factors to DNA.
ACKNOWLEDGEMENTS
We are grateful to Manuel Buchwald (University of Toronto) for the pAS2.1-FANCD2 vector, to Dorina Saro for comments on the manuscript and to Sanja Jahr and Paramjit Uppal for technical help.
FUNDING
National Institutes of Health [RO1CA168635 to P.S and G.M.K, RO1ES015252, PO1CA92584 to P.S., RO1-HL063776 to G.M.K.]; Career Development Award from the Leukemia and Lymphoma Society (5279-08 to S.L.).
Conflict of interest statement None declared.



![DNA substrate specificity in ID2 ubiquitination. (A) ID2 ubiquitination was examined with a variety of oligonucleotide-based DNA substrates as indicated (either 0.4 or 4 μM) or with 4-μM single-stranded oligonucleotides [lanes 12–13 and (B)]. Analysis was by either fluorescent staining (18-h incubation) or western blotting (3-h incubation). Quantification of FANCD2 monoubiquitination after 18 h of incubation is shown. Error bars indicate the standard deviation from at least two independent experiments.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/42/9/10.1093_nar_gku198/1/m_gku198fig3.jpeg?Expires=1716331107&Signature=OK-yBY99LMpcVB87-Nk8b09rdXppQERKzcpX-ZQUEMiRGEa99tOHzFXkkUvqfBuI~jvuFgcVvfiKHxygTQxR6MQGLZAs1xfeP2Lb1NwvEqajSt9Ld0xYzrdVipwHJ~HBFo9Y77Pue2ZnlNKLayNjqkSEP4UkCjfIScxdTyxGu3~Rayp0d4iNxchq0tS~gQakr9rxwdTjSaarnUjhWjZZ3lBwk4sRDpszumNTUl-ROokak1FO5LD0Wg6uZ7FjSG4cZx9YM0ClAicE902FFPc9Gk2OaVn90qkzH7FQyAwJgAJv6h6N2xqHpoxVl58iR0Qx0Rbkgwy6S1HmUddn8wWJ9g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Effects of FANCI mutations on ID2 ubiquitination. (A) DNA binding activity of ID2 complex and mutant variants (harboring the FANCI K294E/K339E, R1285Q or R1285X mutation) was examined as in Figure 4B. Quantification of the data is shown on the right. Error bars indicate the standard deviation of at least two independent experiments. (B) Ubiquitination reactions were carried out with the ID2 complex and its mutant variants with or without DNA [4 μM dsDNA (ds), replication fork DNA (RF) or oligonucleotide O10 (ss)] and analyzed as previously. The incubation time was 18 h for the upper panels and 3 h for the western blots.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/42/9/10.1093_nar_gku198/1/m_gku198fig5.jpeg?Expires=1716331107&Signature=5O6QCpKqwQVMonwXZa-nQ8Cd01M6le8MZBEyLg9MzjDlovIp4oB09Sa7h71DtIPSJM6GeFYbw-tv3TAImChsjmBd2R4MUdh-JwC~nsbmSngTAbnXoLoZuG3Dka34r0yqNKQ5e465h0ZZr2fOszcw8KHv-KdxXLegTtq0DM7FAiDTuuHzZEWxb8SdjrjdkHsQhjAgILtNFBBQqtFr~~WeNL9LQbwTO1TCkh48HyGCrX7mk4gofqHEVji4-cRv1wOCBY6JsnOSjXmZ9XeUj4u6cG1Ky-jdbSnwp762P6VBKN4bKJIeNzJaZ8EDFf1qPScaEEJTxrM39E~2v6-HS2kGQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

Comments