-
PDF
- Split View
-
Views
-
Cite
Cite
Madeleine J. H. van Oppen, Brenda J. McDonald, Bette Willis, David J. Miller, The Evolutionary History of the Coral Genus Acropora (Scleractinia, Cnidaria) Based on a Mitochondrial and a Nuclear Marker: Reticulation, Incomplete Lineage Sorting, or Morphological Convergence?, Molecular Biology and Evolution, Volume 18, Issue 7, July 2001, Pages 1315–1329, https://doi.org/10.1093/oxfordjournals.molbev.a003916
Close - Share Icon Share
Abstract
This study examines molecular relationships across a wide range of species in the mass spawning scleractinian coral genus Acropora. Molecular phylogenies were obtained for 28 species using DNA sequence analyses of two independent markers, a nuclear intron and the mtDNA putative control region. Although the compositions of the major clades in the phylogenies based on these two markers were similar, there were several important differences. This, in combination with the fact that many species were not monophyletic, suggests either that introgressive hybridization is occurring or that lineage sorting is incomplete. The molecular tree topologies bear little similarity to the results of a recent cladistic analysis based on skeletal morphology and are at odds with the fossil record. We hypothesize that these conflicting results may be due to the same morphology having evolved independently more than once in Acropora and/or the occurrence of extensive interspecific hybridization and introgression in combination with morphology being determined by a small number of genes. Our results indicate that many Acropora species belong to a species complex or syngameon and that morphology has little predictive value with regard to syngameon composition. Morphological species in the genus often do not correspond to genetically distinct evolutionary units. Instead, species that differ in timing of gamete release tend to constitute genetically distinct clades.
Introduction
Speciation processes in the sea are expected to differ from those in the terrestrial environment, especially in the case of sessile marine invertebrates. As compared with most terrestrial animals, these organisms display several distinct characteristics in life history and demography that are likely to affect their evolution. For example, they tend to have high dispersal capabilities, large population sizes, wide distributions, extreme longevities—and therefore largely overlapping generations—and high fecundities (Hughes, Ayre, and Connell 1992 ; Palumbi 1994 ). Moreover, many sessile marine invertebrates release their gametes into the water column, where they are mixed by waves and water currents and fertilization takes place. This creates unparalleled opportunities for interspecific hybridization and introgression in species that discharge their eggs and sperm in synchrony with sympatric congeners, as has been documented for many species of reef corals (Harrison et al. 1984 ; Willis et al. 1985 ; Babcock et al. 1986 ). Experimental breeding trials with scleractinian corals confirm that reticulate pathways are likely to have contributed to the evolution of modern Indo-Pacific coral species in the genera Acropora, Montipora (Willis et al. 1997 ), and Platygyra (Miller and Babcock 1997 ). Ribosomal DNA internal transcribed spacer (rDNA ITS) sequence (Odorico and Miller 1997 ), mini-collagen second intron (Hatta et al. 1999 ), and allozyme (Miller and Benzie 1997 ) results are consistent with these findings. Similar patterns have also been observed in Caribbean corals belonging to the genera Acropora (van Oppen et al. 2000 ) and Madracis (Diekmann et al. 2001 ).
Interspecific hybridization can lead to a mosaic composition of the nuclear genome due to introgression and recombination. Cytoplasmic genomes, however, usually show clonal transmission and are predominantly maternally inherited. This allows for an accurate identification of ancestral sources. Moreover, the comparison of phylogenies based on nuclear (nDNA) versus cytoplasmic DNA markers should make it possible to identify cases of hybridization by focusing on the discrepancies between them (e.g., Avise 1994 and references therein; Parani et al. 1997 ). In Helianthus sunflowers, for example, the chloroplast DNA (cpDNA)–based phylogeny contrasts significantly with implied relationships based on morphological characters and nuclear ribosomal DNA (Rieseberg and Soltis 1991 ). Each of 5 species (of the 11 species investigated) possesses two or more highly distinct cpDNA genotypes otherwise characteristic of different phylogenetic groups within the genus. Furthermore, some species seem to have captured the cpDNA of other species on multiple occasions. Several similar examples exist from animal systems, in which mitochondrial DNA (mtDNA) capture was found to be due to introgression among closely related species (e.g., Lamb and Avise 1986 ; Arnason et al. 1991 ; Dowling and Hoeh 1991 ; Dowling and DeMarais 1993 ). In the cyprinid fish genus Gila, for instance, comparison of allozyme and mtDNA data showed that introgression not only has occurred in the past, but is still proceeding today (Dowling and DeMarais 1993 ).
The genus Acropora (Scleractinia, Acroporidae) is one of the most widespread genera of corals, spanning the Indian and Pacific Oceans and the Caribbean Sea. It is also extremely speciose and the largest extant reef-building coral genus. Recent revisions of the genus recognize 113 (Wallace 1999 ) or approximately 180 (Veron 2000 ) Acropora species. The genus Acropora consists of two subgenera, A. Acropora and A. Isopora (Veron and Wallace 1984 ), with 19 Indo-Pacific and 1 Caribbean species group being recognized on the basis of skeletal morphology within the former subgenus (Wallace 1999 ). Some species have very restricted distributions, whereas others are found throughout large parts of the tropics, and up to 70 Acropora species can be found in sympatry (Veron 1993 ). An enormous amount of intraspecific morphological variability exists, while at the same time similarities between species are striking; for example, intraspecific geographic differences in morphology can be as large as differences between species (J. E. N. Veron, personal communication). The fossil record shows that the genus probably originated during the Paleocene (Carbone et al. 1994 ) or the Eocene (von Fritsch 1895; Latham 1929 ) and became widely distributed in the early Miocene (Wells 1956 ). Acropora thus provides an ideal model system for examining speciation and evolution of scleractinian reef coral species in general, on both temporal and spatial scales.
Here, we present molecular phylogenies of a wide range of species in the subgenus A. Acropora using DNA sequence analyses of two molecular markers, one nuclear and one mitochondrial. The nuclear intron was the Pax-C 46/47 intron. Acropora Pax-C is a Pax-6 homolog (Catmull et al. 1998 ), and in almost every organism that has been studied, these are single-copy genes (Callaerts, Halder, and Gehring 1997 ). The mitochondrial region examined here is the largest intergenic region identified so far in the Acropora tenuis mitochondrial genome and has several characteristics of a vertebrate control region (van Oppen et al. 1999 ). The molecular phylogenies are compared with cladistic analyses of morphological features of the skeleton (Wallace 1999 ) and discussed in the context of the fossil record, taxonomic uncertainty, lineage sorting, and interspecific hybridization.
Materials and Methods
Sample Collection for DNA Analysis
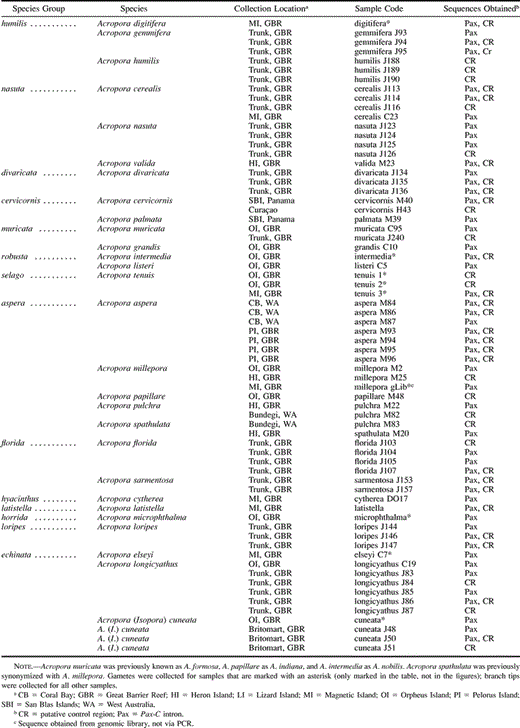
Tissue samples were collected by snapping off small (2–5-cm) branches from individual colonies and were stored in 90%–100% EtOH, which was replaced after 1–2 weeks. Sperm and eggs were collected during the annual coral mass spawning event and frozen in liquid nitrogen. Most samples were collected in the central Great Barrier Reef (GBR) region, Eastern Australia: Magnetic Island, Orpheus Island, Pelorus Island, Britomart, and Trunk Reef. In addition, a small number of colonies were collected from Lizard Island (northern GBR), Heron Island (southern GBR), Western Australia (Coral Bay and Bundegi), and the Caribbean (Acropora palmata and Acropora cervicornis) (table 1 ).
DNA Extraction, PCR Conditions, and Cloning and Sequencing Procedures
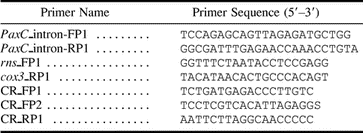
DNA was extracted from approximately 1 cm3 of coral branch as described in van Oppen, Willis, and Miller (1999) . PCR amplification was carried out using the following primer pairs and conditions: (1) For the Pax-C intron, PaxC_intron-FP1 and PaxC_intron-RP1 (table 2 ) were used. The forward primer was located 112–90 bp upstream of the intron. The reverse primer annealed to the 3′ end of the intron (last 5 nt) and the 3′ coding in a region of high amino acid identity. These primers were also used for sequencing. One microliter of 10× or 50× diluted DNA was added to 24 μl of PCR master mix consisting of 1 μl of each primer at a concentration of 10 μM, 2.5 μl dNTPs (2 mM), 2.5 μl 10 × reaction buffer (Fisher Biotech), 2 μl MgCl2 (25 mM), 0.13 μl Taq polymerase (5.5 U/μl; Fisher Biotech), and 14.87 μl H2O. The PCR profile consisted of an initial denaturation step of 3 min at 95°C, followed by 5 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C, followed by 26 cycles as before but with an annealing temperature of 56°C instead of 50°C. Finally, the mixture was incubated at 72°C for 10 min. (2) For the mtDNA putative control region (hereinafter referred to as the mtDNA intergenic region), PCR primers were designed from the A. tenuis mtDNA rns and cox3 sequence (van Oppen et al. 1999 ) (table 2 ), which flank the mtDNA intergenic region (rns_FP1 and cox3_RP1). Using these primers and the PCR cocktail as before, the mtDNA intergenic region was amplified using an initial denaturation step of 30 s at 94°C, followed by 30 cycles of 20 s at 94°C, 30 s at 54°C, and 90 s at 72°C. Finally, the mixture was incubated at 72°C for 5 min. The PCR, as well as additional internal primers (CRFP1, CRFP2, and CRRP1; table 2 ) and the vector primers T7 and SP6, were used for sequencing.
PCR products were run on a 0.8% TAE-agarose gel, excised, and purified using the Jetsorb (Genomed) kit. DNA concentration was measured with a spectrophotometer at 260 nm, and the products were cloned in pGEM-T (Promega) following the manufacturer's instructions. Positive clones were either amplified using colony PCR and purified for sequencing as described in van Oppen, Willis, and Miller (1999) or grown overnight in LB medium followed by a Qiagen QIAprep spin plasmid miniprep according to the manufacturer's instructions. Sixty to ninety nanograms of PCR product or 200 ng of plasmid DNA was used for a 10-μl sequencing reaction consisting of 1 μl primer at a concentration of 3.3 μM, 4 μl of Perkin Elmer BigDye terminator mix (PE Applied Biosystems), and 5 μl of DNA solution. Sequencing products were separated on an ABI 310 Genetic Analyzer.
For the Pax-C intron, 2–7 clones were initially sequenced from a subset of coral colonies to verify that Pax-C was single-copy. Most coral colonies possessed only one or two different sequences. In those few instances in which more than two sequences per individual were found, these differed by only one or two point mutations which were not shared by other clones, suggesting that these were PCR errors and that no more than two alleles were present per diploid genome. After this initial screening, only two clones were sequenced per coral colony (one in both directions, one in only one direction). If the two sequences were identical, two more clones were sequenced to investigate whether the colony was heterozygous for Pax-C or not. For the mtDNA intergenic region, only one clone was sequenced for each coral colony.
Data Analyses
Sequences were aligned manually using Sequencher 3.0 (Gene Codes Corporation). Maximum-likelihood (ML) analyses were performed in MOLPHY 2.3 (Adachi and Hasegawa 1996 ), under the HKY85 model of sequence evolution. Analyses under a constraint topology were carried out in MOLPHY 2.3 and in PAUP* 4.0b2a (Swofford 1999 ) to examine statistical significance of the difference in tree topologies using the Kishino-Hasegawa test (Kishino and Hasegawa 1989 ). Neighbor-joining distance and parsimony (heuristic search) analyses were conducted in PAUP*. Gaps were either excluded or treated as fifth bases in parsimony analyses. Bootstrap analysis was performed with 1,000 replicates whenever possible, but with fewer replicates if computing time became excessive. Pairwise sequence distances were calculated in PAUP* 4.0b2a.
The Pax-C intron alignment contained several large indels. Parsimony analyses were performed with some of the large indels treated as single alignment positions with two character states (present/absent). To minimize loss of information, only indels where no or few informative sites were present among taxa that had an insert were removed from the alignment (and treated as single sites). Four Pax-C intron indels of 529, 403, 42, and 145 bp (in total, 1,119 positions) were excluded and recoded. These new positions were weighted according to their sizes, as follows: 47 ((529/1,119) × 100), 36, 4, and 13, respectively. The analyses were also carried out by assigning arbitrary weights of 10 or 25 to the recoded indel positions. The distance analyses were performed on the original alignment, with gaps treated as missing data. The mtDNA intergenic region contained several repeated sequences (the Acropora mtDNA intergenic region structure is discussed in detail elsewhere [van Oppen et al., unpublished data]). Since the precise processes by which these repeat arrays evolve are not well understood (Hoelzel, Hancock, and Dover 1993 ; Broughton and Dowling 1994 ; Fumagalli et al. 1996 ), and hence their phylogenetic information content is dubious, phylogenetic analyses were performed with and without these regions.
Trees obtained from each of the two data sets (i.e., the Pax-C intron and the mtDNA intergenic region) were compared using the Kishino-Hasegawa test in PAUP* 4.0b2a.
Results
Base Composition, Genetic Distances, and Saturation
Base compositions of the nuclear and mitochondrial DNA regions is given in table 3 . The Pax-C intron had a bias toward A and T and away from G, which is quite different from the base composition found for Pax-C exons from Acropora millepora by Catmull et al. (1998) (i.e., 0.323 [A], 0.214 [C], 0.228 [G], and 0.232 [T]). Base frequencies differed considerably between the two subgenera. The A. Isopora nuclear sequences had a lower T content and a higher C content compared with those of A. Acropora. It should be noted that sample sizes for A. Isopora were relatively small. The mtDNA control had a bias toward T and strongly away from C. AT content in this region (both repeat and nonrepeat regions combined) was 57.4 %, slightly lower than the average AT percentage of the mtDNA coding and intergenic sequences of A. tenuis (62.1%; van Oppen et al., unpublished data). No significant difference in base frequencies was observed between A. Acropora and A. Isopora mtDNA intergenic region sequences.
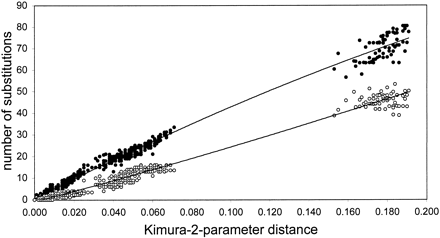
Maximum corrected pairwise sequence distances within the subgenus A. Acropora were higher for the nuclear intron than for the mtDNA intergenic region (table 4 ). In the comparison between subgenera, however, maximum sequence differences were similar for the Pax-C intron and the mtDNA intergenic region. These results may be somewhat biased due to the small sample size for the A. isopora sequences. Nevertheless, for both markers, sequence distances were largest between the two subgenera A. Acropora and A. Isopora. The asymptotic transition/transversion (ts/tv) ratio (indicating saturation for base changes) is dependent on base frequencies (Holmquist 1983 ) and is 0.48–0.49 for the Pax-C intron. In all comparisons (both within and between the two subgenera), most ts/tv ratios were >1, and only a very small fraction of them were found to be <0.48. Some of the values <0.48 occurred in comparisons of very similar sequences, suggesting that these values were statistically unreliable due to the small number of base changes sampled. Hence, there was no evidence for saturation in the Pax-C intron sequence comparisons presented here. Mitochondrial sequences often have a great excess of transitions over transversions. To assess levels of sequence saturation in the mtDNA intergenic region, numbers of transitions and transversions were plotted against corrected (Kimura two-parameter) pairwise sequence differences (fig. 1 ). The curve of transitions against sequence divergence revealed only a slight trend toward asymptotic saturation among the most divergent taxa (A. Isopora vs. A. Acropora), and no evidence for saturation of transversions was observed. Approximately twice as many transitions as transversions have occurred among A. Acropora sequences. Transversions were therefore weighted twice as heavily as transitions in some of the parsimony analyses (see below).
Phylogenetic Analyses
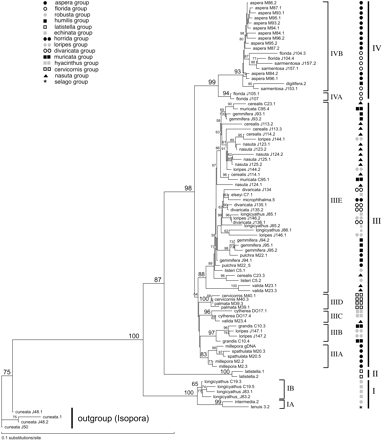
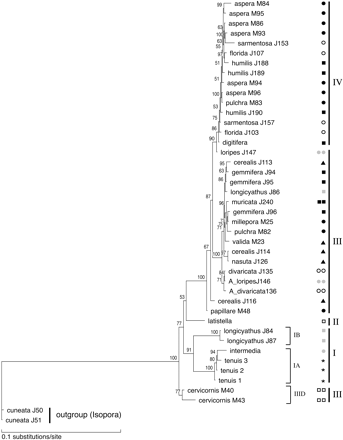
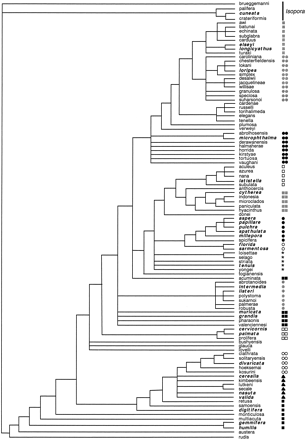
Figure 2A and B shows the ML trees based on the nuclear and mitochondrial markers, respectively. Trees were rooted using Acropora (Isopora) cuneata because the subgenus A. Isopora forms the sister group to A. Acropora in cladistic analyses using morphological characters of the skeleton (Wallace 1999 ; fig. 3 ). Sequence analysis of Cytochrome b and NADH dehydrogenase subunit 2 confirm that A. Isopora is the sister group to A. Acropora (van Oppen, Willis, and Miller 1999 ; Fukami, Omori, and Hatta 2000 ). Below, the results of the phylogenetic analyses are discussed in detail for each of the two markers separately.
Pax-C Intron
Of the 45 individuals for which DNA was extracted from tissue (five DNA extracts were from gametes, and one sequence came from a genomic library), 33 were heterozygous for the Pax-C intron (Ho = 0.73). This is a minimum estimate because of the way in which individuals were screened for heterozygosity (see above). We are currently screening many individuals for variation at this intron using SSCP analysis, which should provide more accurate estimates of heterozygosity. The only species for which we have found more than two alleles is Acropora valida, which is known to be polyploid (Kenyon 1997 ). Of the 22 species of Acropora studied by Kenyon (1997) , there are three possible cases of polyploidy. Of the species included in our study, Acropora elseyi may be tetraploid and A. valida triploid. The A. elseyi DNA sample used in our study was extracted from gametes. Hence, we could have observed a maximum of two alleles. The fact that we observed only a single allele may be due to the individual having been homozygous or to the small number of clones (i.e., two clones) sequenced. The mean chromosome numbers for Acropora divaricata and Acropora gemmifera reported by Kenyon (1997) were 30, compared with 28 in most Acropora (and other coral) species. These numbers do not imply polyploidy. In concordance with this, we have not found more than two alleles in any other species (this has been confirmed by SSCP analysis), supporting the notion that Pax-C is single-copy in Acropora. Although alleles within individuals showed considerable sequence divergence in many cases, they did fall within the same major phylogenetic clade (see fig. 2A ).
The length of the sequenced region of the intron (the last 32 bases at the 3′ end were excluded because this sequence had not been obtained reliably for all taxa) varied from 212 to 827 bp (GenBank accession numbers AF344337–AF344423). The complete Pax-C intron sequence alignment consisted of 1,613 positions, of which 1,381 were constant, 100 were variable but not parsimony-informative, and 132 were parsimony-informative (when gaps were treated as missing data). In some analyses, a total of 1,119 positions (comprising four large indels) were excluded and recoded as four new alignment positions (see Materials and Methods). This modified alignment consisted of 494 positions, of which 96 were constant, 97 were variable but parsimony-uninformative, and 301 were parsimony-informative (gaps treated as fifth bases). Phylogenetic signal in this modified data set was high, as indicated by relatively high consistency (CI) and retention (RI) indices (CI = 0.714–0.733 and RI = 0.852–0.872, depending on which weight was given to the recoded indels) of the resulting trees and a significantly left-skewed tree length distribution, which was tested by generating 10,000 random trees (g1 = −0.602) (Hillis and Huelsenbeck 1992 ).
Pax-C Intron Phylogeny
Parsimony analysis using the modified alignment with gaps treated as fifth bases yielded 240 most-parsimonious trees (MPTs). The overall topology of the parsimony trees did not differ with different weighting of the four indels and identified four major clades within the A. Acropora subgenus. Neighbor-joining distance and ML analyses yielded trees similar to those of the parsimony analyses (fig. 2A ). The two most basal clades, clades I and II, were small, comprising three (Acropora longicyathus, Acropora intermedia, and A. tenuis) and one (Acropora latistella) species, respectively. The third clade was much larger and comprised 18 species in many well-supported terminal clades—including the Caribbean species (A. cervicornis and A. palmata)—but showed limited resolution at its base. The fourth clade comprised two main subclades and only four species (Acropora aspera, Acropora florida, Acropora sarmentosa, and Acropora digitifera). All colonies of each species consistently clustered in the same major clade except for four colonies of A. longicyathus, which were split between clades II and III. Despite this, many species were not monophyletic. We tested whether species were truly nonmonophyletic (for those of which more than one colony was sampled) by enforcing species monophyly (one species at a time, as well as all species at a time) and comparing the resulting tree with the best tree (fig. 2A ). In 7 of the 10 species tested, a significantly worse tree was obtained by enforcing monophyly (table 4 ), indicating that these species were not monophyletic. The most striking case of polyphyly was that of A. longicyathus, for which some alleles/haplotypes clustered in the basal clade I and some in the derived clade III. Note that the sample sizes were small for some species and that polyphyletic patterns like the one observed for A. longicyathus may possibly be found in other species with the addition of new samples. Trees that were forced to follow the topology based on morphological characters (Wallace 1999 ; fig. 3 ) were also significantly worse than the best tree (−ln L = 5,917.6 for the null hypothesis of monophyly of species and species groups, as compared with −ln L = 4,457.1 for that of the optimal tree, P < 0.0001), indicating that morphological species groups do not constitute natural groups.
mtDNA Intergenic Region
The complete mtDNA intergenic region alignment (both repeat and nonrepeat regions) consisted of 1,726 positions. This included nine bases of the upstream small subunit rRNA (rns), because the first repeat region started within this gene. The length of the whole mtDNA intergenic region (excluding the nine bases of the rns) varied between 999 and 1,222 bp (GenBank accession numbers AY026418–AY026460). No size heteroplasmy was observed by PCR. As we sequenced only one clone per coral colony, small intraindividual length differences in the mtDNA intergenic region may have been overlooked. Because there was a much stronger phylogenetic structure in the data set when repeat regions were excluded, only this data set is discussed here. The alignment of nonrepeat regions consisted of 816 positions, of which 657 were constant, 47 were variable but parsimony-uninformative, and 112 were parsimony-informative (gaps treated as fifth bases). Treating gaps as missing or fifth bases did not lead to an altered tree topology. Parsimony analysis with gaps treated as fifth bases yielded 240 MPTs of 251 steps. A CI of 0.834, an RI of 0.921, and a g1 value of −0.865 are indicative of a strong phylogenetic signal in the data set. The analysis was also run while weighting transversions more heavily than transitions (1:0, 2:1 [within A. Acropora, approximately twice as many transitions as transversions have occurred; fig. 1] , and 10:1). The resulting tree topologies were very similar to each other and to the ones without ts/tv weighting.
mtDNA Intergenic Region Phylogeny
Neighbor-joining distance analyses produced tree topologies almost identical to those of the parsimony analyses, but with slightly more resolution in some of the terminal clades. ML analysis (HKY85 model) also yielded a similar tree but had the highest level of resolution (fig. 2B ). Bootstrap analysis showed that several robust clades could be distinguished. Two large and three relatively small clades were identified. Two of the small clades had identical species compositions, as was found with the nuclear marker (i.e., one clade was composed of A. longicyathus, A. intermedia, and A. tenuis, with the former species clustering separately from the latter two, and a second clade was composed solely of A. latistella). Again, different colonies of A. longicyathus clustered separately in two of the clades. However, the Caribbean species A. cervicornis formed a separate and most basal clade in the tree based on the mtDNA intergenic region, in contrast to its inclusion in the large clade in the Pax-C intron tree.
Congruent Phylogenetic Patterns Based on a Nuclear and a Mitochondrial Marker?
Congruence between trees based on the Pax-C intron and the mtDNA intergenic region was assessed by the Kishino-Hasegawa test (Kishino and Hasegawa 1989 ). Because a requirement of this test is that both data sets are derived from the same individuals, it was performed on the subset of samples (25 specimens) for which both regions were successfully amplified and sequenced (see table 1 ). For these 25 specimens, the ML trees were first calculated based on the two markers independently, after which the Kishino-Hasegawa test was applied reciprocally (i.e., using each data set at a time) under the ML criterion. The trees were significantly different (P ≤ 0.0003 in all cases). Nevertheless, visual inspection of the trees based on the full data sets shows that the overall compositions of the four major clades are similar between the two trees. In both trees, (1) A. tenuis, clade IB samples of A. longicyathus, and A. intermedia form the basal taxa; (2) A. latistella is the next species to split off; (3) the remaining species fall into two clusters; and (4) these two clusters largely share species; for example, A. aspera, A. sarmentosa, A. florida, and A. digitifera occur in clade IV of both the mtDNA and the nDNA trees. However, there are also discrepancies between the trees: (1) A. loripes J147 falls within clade IV based on mtDNA, while it clusters in clade IIIB based on the Pax-C data, and (2) the Caribbean species form the most basal clade in the mtDNA tree but constitute a derived clade (IIID) based on the Pax-C data. In addition, one A. pulchra specimen (pulchra_M83) falls within clade IV in the mtDNA tree. Although no Pax-C sequences are available for this individual, rDNA ITS sequences have shown that it is likely to be a hybrid between A. aspera and A. pulchra (unpublished data), explaining its position in the mtDNA tree.
Discussion
Introgressive Hybridization or Incomplete Lineage Sorting?
The incongruence between the nuclear and the mitochondrial trees, in combination with the nonmonophyly of many species, suggests either that introgressive hybridization has occurred or that lineage sorting is incomplete. If species are of recent origin, the latter explanation is likely to be at least partly responsible. Unfortunately, the ages of most species are not known due to the scarcity of Acropora fossils that can be reliably identified to the species level in the Indo-Pacific. Due to the fourfold smaller effective population size of mtDNA compared with nDNA, lineage sorting is expected to reach completion much faster in mtDNA. Hence, if the differences between the two trees are truly caused by differences in the extent of lineage sorting, the mtDNA tree is likely to represent the species tree most accurately. This would suggest that the Caribbean Acropora species are most ancestral. Possibly, some of the polytomies in the derived clades III and IV may represent explosive speciation of their Indo-Pacific descendants during the Pliocene and the Pleistocene (see below). Hence, it is difficult to reject the possibility that incomplete lineage sorting is responsible for the observed patterns. However, two pieces of additional independent evidence lead us to favor the hybridization hypothesis. First, many species are capable of successful cross-fertilization with a range of congeners (Willis et al. 1997 ), and second, species forming genetically distinct clusters in the phylogenetic analyses tend to differ with regard to the time of gamete release, as discussed next.
Timing of Spawning, Reproductive Isolation, and Genetic Distinctness
Independent evidence from timing of spawning provides support for evolutionary relationships detected by the molecular analyses. Several of the basal species in the molecular phylogenies appear to be reproductively isolated from most other species in the A. Acropora subgenus through differences in spawning times. Acropora latistella typically spawns two weeks out of phase with most A. Acropora species (Willis et al. 1985 ; Babcock et al. 1986 ), supporting its isolated position in the molecular phylogenies. On the Great Barrier Reef, A. tenuis is likely to be effectively genetically isolated by a temporal separation in breeding, since it spawns 2–3 h earlier than its congeners (Babcock et al. 1986 ). Although eggs and sperm may remain viable for 4.5–8 h after spawning (Heyward and Babcock 1986 ; Oliver and Babcock 1992 ; Willis et al. 1997 ), it is unlikely that eggs would remain unfertilized or sperm densities would remain high enough in the intervening period to achieve high fertilization rates with later-spawning species. In the Caribbean, a difference in spawning time of 1–2 h appears to be sufficient to cause reproductive isolation between some species in the Montastrea annularis complex (Knowlton et al. 1997 ). Moreover, A. tenuis displayed very low breeding compatibilities with other species in vitro (Willis et al. 1997 ), also confirming its separate species status. In addition to differences in spawning times, transient reproductive barriers imposed by year-to-year variation in the date on which spawning occurs may also contribute to the relative genetic distinction of Acropora species.
Acropora aspera is genetically distinct from other species in the A. aspera group (e.g., A. millepora, A. spathulata, A. papillare, and A. pulchra—with the exception of F1A. aspera × A. pulchra hybrids) in our analyses, as well as in an analysis of rDNA ITS regions (unpublished data), and has been observed to spawn one month before most of its congeners in some years (unpublished data). The exception of F1 A. aspera × A. pulchra hybrids is consistent with evidence that these two species are capable of successful cross-fertilization in years when they spawn synchronously (unpublished data). Note that a Pax-C tree in which monophyly of A. aspera was enforced was not significantly worse than the tree shown in figure 2A (table 4 ). Acropora intermedia also shows some temporal variation in the date of spawning and has spawned before other congener species around the Palm Islands in the central Great Barrier Reef in some years (unpublished data), suggesting that, at least in some years, it may be reproductively isolated through differences in the timing of spawning. It also displayed very low breeding compatibilities with other species in vitro (Willis et al. 1997 ).
The phylogenetic isolation of species with different spawning times and limited cross-fertility provides evidence that hybridization is likely to be responsible for the nonmonophyly of sympatric species which spawn synchronously. Of the 21 Indo-Pacific species that group in the two large clusters, all have been recorded to spawn on the same night as 2–12 other species in these clades (Harrison et al. 1984 ; Willis et al. 1985 ; Babcock et al. 1986 ), and 5 have been shown to successfully hybridize in vitro with one of these congeners (Willis et al. 1997 ). Although A. nasuta, A. valida, and A. muricata spawn synchronously, they had very limited interspecific breeding compatibilities (Willis et al. 1997 ). However, only a limited number of colony pairs were tested for these species (Willis et al. 1997 ), and given the enormous variability in gamete compatibility in most interspecies pairings (Willis et al. 1997 ), the low interspecific breeding compatibility indicated for these species may be an artifact of small sample size. Another possible explanation for this apparent discrepancy is that indirect introgression may occur in nature—alleles may be exchanged between species that do not directly hybridize, via a third species with which both hybridize. Alternatively, some of these species may have become reproductively isolated in the recent past, and sharing of alleles may reflect shared ancestral polymorphisms.
Molecular Data Are Inconsistent with Morphology and the Fossil Record
With the exception of the position of the Caribbean taxa and A. loripes J147, the compositions of the four major clades in phylogenetic trees based on a nuclear and a mitochondrial marker were similar, albeit the within cluster topologies varied (fig. 2A and B ). However, these phylogenies bear little similarity to a recent cladistic phylogeny based on skeletal morphology (fig. 3 ; Wallace 1999 ) and are at odds with the fossil record. Although the record for extant A. Acropora species is scant before the Plio-Pleistocene, two species (Acropora humilis and Acropora microphthalma) have been recorded from the Miocene (Wells 1964 ). Both species would therefore be expected to have a basal position in the phylogenetic trees. However, rather than constituting a basal clade, members of the A. humilis group, as well as A. microphthalma, are scattered through different subgroups of the derived clades III and IV. Note that using midpoint (rather than outgroup) rooting did not affect the derived positions of these species (data not shown).
These inconsistencies between the different data sets can be explained in two ways. First, it is possible that different morphologies have independently evolved more than once, and second, skeletal morphology may have been effectively uncoupled from the genotype in the case of Acropora evolution. These hypotheses are not mutually exclusive and would be difficult to distinguish—the main difference between them is that the second is hybridization-dependent, whereas the first is not. The observation that presumed F1 hybrids between A. aspera and A. pulchra have morphological features of one of the parental species rather than an intermediate morphology (unpublished data) suggests that morphology is defined by a limited number of (closely linked) genes with dominant/recessive rather than codominant alleles. If this is generally true, then the particular combination of alleles determining, for example, the A. humilis morphology may have arisen several times independently; alternatively, that combination of alleles may have been transposed into different genetic backgrounds via introgressive hybridization. The differences between the mitochondrial and nuclear DNA phylogenies imply that introgressive hybridization may be occurring. However, the overall congruence between the four major clades suggests that hybridization between species from different main clades is limited. Many A. Acropora species are capable of successful interspecific hybridization in vitro (Willis et al. 1997 ; Hatta et al. 1999 ), and genetic data confirm that introgressive hybridization occurs in nature (Odorico and Miller 1997 ; Hatta et al. 1999 ; van Oppen et al. 2000 ). Hence, the second hypothesis is the most plausible. Although no breeding trials have been performed using A. humilis or A. microphthalma, it is possible that these species are reproductively compatible with several other A. Acropora species.
The Effect of Major Vicariant Events on the Evolution of A. Acropora Species
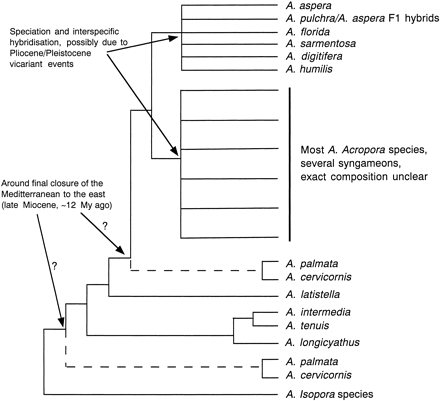
Although no intraspecific biogeographic structure is obvious in the genetic data, we might expect to be able to detect the influence of major vicariant events on the evolution of the genus. Figure 4 summarizes a phylogenetic hypothesis in which the impact of geological and climatological events is superimposed onto a phylogenetic tree based on the sequence analyses presented in this study. The main difference among the nuclear and mitochondrial trees is the position of the Caribbean taxa, which are the most ancestral A. Acropora species in the mtDNA tree but are part of the large clade III in the Pax-C intron tree (fig. 2A and B ). As discussed before, this incongruence is possibly due to incomplete lineage sorting in one or both markers. The node from which the Caribbean species emerge is likely to coincide with the effective isolation between Caribbean and Indo-Pacific A. Acropora species. The earliest A. cervicornis and A. palmata fossils are approximately 6.6 Myr old (Budd and Johnson 1999 ) and 3.6–2.6 Myr old (McNeill, Budd, and Borne 1997 ), respectively. The third extant Caribbean Acropora species—which was not included in this study but has been found to have Pax-C intron sequences similar to those of A. palmata and A. cervicornis (and the three species constitute a monophyletic group) (van Oppen et al. 2000 )—is of Holocene origin (Budd, Stemann, and Johnson 1994 ). Hence, the internode denoting the emergence of the Caribbean species must represent a point in time before 6.6 MYA, which was well before the closure of the Isthmus of Panama (3.0–3.5 MYA; Keigwin 1982 ; Duque-Caro 1990 ; Coates and Obando 1996 ). The Tethys Sea remained an open east-west connection between the present-day Atlantic and Pacific Oceans via the Mediterranean until approximately the mid-Miocene (ca. 12 MYA; Rögl and Steiniger 1984 ; Por 1989 ), although it seems likely that there were several closures and reopenings from the late Oligocene or early Miocene (Rosen 1988 ). The latest possible date of contact between the Caribbean and Indo-Pacific taxa via the Middle East is approximately 12 MYA at the time of the final closure of the Tethys Sea. However, as with biota on both sides of the Isthmus of Panama (Knowlton et al. 1993 ), the differentiation of Caribbean and Indo-West Pacific A. Acropora species may have preceded the actual closure of the Tethys Sea in the Middle East. If the Caribbean A. Acropora species really represent the most ancestral extant species in the genus, then this implies that most Indo-Pacific Acropora species have evolved over the last 10 Myr or so.
While there are several robust terminal clades present within clades III and IV, there are also many unresolved nodes within these clades (figs. 2A and 4 ). The level of genetic divergence among taxa suggests that these unresolved nodes reflect true polytomous events (hard polytomies; Hoelzer and Melnick 1994 ) rather than a failure of the marker (the Pax-C intron) to resolve this node. Hard polytomies may reflect explosive speciation events, simultaneous fragmentation of populations, or introgressive hybridization and recombination events. In the case of A. Acropora, it might well be a combination of all three possible causes. A major sea-level regression took place toward the end of the Miocene/early Pliocene (Vail and Hardenbol 1979 ). A series of sea-level fluctuations also occurred in the Pleistocene (Haq, Hardenbol, and Vail 1987 ; Chappell et al. 1996 ), and some coral reef organisms, such as butterfly fishes (McMillan and Palumbi 1995 ) and starfish (Benzie 1999 ), are believed to have undergone a diversification as a result of this. These sea level changes would have led to changes in intensity and direction of sea surface currents and, as a consequence, would have caused the repeated isolation and (re)connection of populations and species (Veron 1995 ; Benzie 1999 ). As sea levels rose, new habitats became available, and a combination of founder effects and selection may have caused many new species to evolve (Potts 1983 ). The collision of Australia/New Guinea with the islands off the Asian continent, which began ca. 15 MYA (Audley-Charles 1981 ; Hall and Holloway 1998 ), may have further contributed to this period of evolutionary diversification.
The molecular phylogenies presented here imply that many reef corals undergo reticulate evolution, as predicted by Veron (1995) . Thus, clusters that comprise several morphospecies represent a syngameon, i.e., “the sum total of species or semispecies linked by frequent or occasional hybridization in nature; [hence] a hybridizing group of species …” (Grant 1957 ). A syngameon may consist of a large number of species (corresponding to morphospecies), each with varying levels of reproductive compatibilities with every other species. For example, the cluster IV comprising A. aspera, A. florida, A. sarmentosa, A. digitifera, and A. humilis may constitute a syngameon. Moreover, since A. aspera and A. pulchra are known to occasionally produce viable offspring, clades III and IV would represent a single syngameon according to the definition given above. Alternatively, morphospecies in clades III and IV could be incipient species that are still partly connected through occasional crossings. The molecular data presented here cannot distinguish between these two hypotheses, but a comparison with recently published molecular data on Acropora species off Japan support the reticulate-evolution hypothesis (see below).
Biogeographic Variation in the Composition of Syngameons
Hatta et al. (1999) performed experimental breeding trials and DNA sequence analysis of the mini collagen second intron using A. nasuta, A. digitifera, A. muricata (=A. formosa), A. florida, and A. intermedia (=A. nobilis) from Okinawa, Japan. The results of this study are not concordant with our work; when we forced our data to follow their tree, the resulting ML tree was considerably worse (−ln L equals; 1,601.8, as compared with 1,467.6 for our optimal tree). In the Hatta et al. (1999) tree, A. florida and A. intermedia were found to be genetically closely related, whereas in our study, A. florida and A. intermedia cluster in two of the most distinct clades of the tree (i.e., clades IV and I, respectively). A second clade in the Hatta et al. (1999) study comprised A. digitifera, A. nasuta, and one morph of A. muricata. In contrast, our study suggests that although A. nasuta and A. muricata carry relatively closely related genomes, A. digitifera should be placed in a different subclade. The differences between these two studies may partly be related to sample sizes. These are small for some of the species (e.g., A. intermedia) investigated in our study. In the Hatta et al. (1999) study, only one pair of colonies was crossed for each of the species/morph combinations, despite the fact that fertilization rates may vary widely in experimental crosses (Willis et al. 1997 ). Alternatively, true differences may exist in the degree to which species hybridize in different biogeographic regions. Acroporid species composition in Japan differs from that on the GBR, as indicated by a Jaccard's similarity coefficient of 0.78 (Wallace 1999 ), and this may affect patterns of interspecific hybridization in the two biogeographic regions.
Conclusions
Based on the molecular data presented here, we hypothesize that the subgenus A. Acropora is composed of several species complexes or syngameons, each consisting of a range of morphospecies, and that many morphological species in this genus do not correspond to genetically distinct evolutionary units. Although alternative explanations, such as shared ancestral polymorphism causing species poly- and paraphyly in phylogenetic analyses of molecular data, cannot be refuted at this time, previous studies on cross-fertility and spawning times of a range of Acropora species support our hybridization hypothesis.
Stephen Palumbi, Reviewing Editor
Present address: Tropical Environment Studies and Geography, James Cook University, Townsville, Australia.
Keywords: reticulate evolution interspecific hybridization Acropora coral mtDNA nuclear DNA
Address for correspondence and reprints: Madeleine J. H. van Oppen, Biochemistry and Molecular Biology, James Cook University, Townsville 4811, Australia. E-mail: madeleine.vanoppen@jcu.edu.au.
Table 3 Mean Base Compositions (%) of the Pax-C Intron and the mtDNA Intergenic Region (both repeat and nonrepeat regions) in Acropora Species

Table 3 Mean Base Compositions (%) of the Pax-C Intron and the mtDNA Intergenic Region (both repeat and nonrepeat regions) in Acropora Species

Table 4 Ranges of Kimura Two-Parameter Pairwise Sequence Distances (%) for the Pax-C Intron and the mtDNA Intergenic Region (nonrepeat regions only) Among and Within Acropora Acropora Species and Between A. Acropora and Acropora Isopora Species

Table 4 Ranges of Kimura Two-Parameter Pairwise Sequence Distances (%) for the Pax-C Intron and the mtDNA Intergenic Region (nonrepeat regions only) Among and Within Acropora Acropora Species and Between A. Acropora and Acropora Isopora Species

Table 5 Results of Kishino-Hasegawa Tests of Species Monophyly for the Pax-C Intron Data
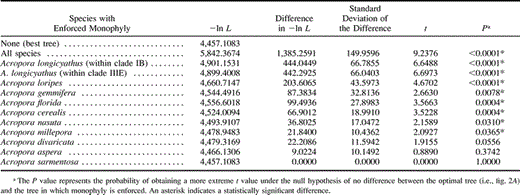
Table 5 Results of Kishino-Hasegawa Tests of Species Monophyly for the Pax-C Intron Data

Fig. 1.—Apparent number of transitions (filled circles) and transversions (empty circles) plotted against corrected (Kimura two-parameter) pairwise sequence difference for the mtDNA intergenic region (nonrepeat regions only)
Fig. 2.—Rooted maximum-likelihood trees using the HKY85 model of sequence evolution for (A) the Pax-C intron and (B) the mtDNA intergenic region. Values on the branches indicate bootstrap values (1,000 replicates). Symbols for the different species groups are given in A. The four major clades are indicated by I, II, III, and IV. Subclades within these clades are marked by a letter followed by a number (e.g., IIIa). Sample codes are as in table 1 . A dot and a number after the sample code indicates the clone that was sequenced (e.g., cerealis J113.2 = Acropora cerealis colony J113, clone 2)
Fig. 2 (Continued)
Fig. 3.—Parsimony consensus tree based on 23 morphological characters of the skeleton (redrawn from Wallace 1999 ). Species that were used for the molecular phylogenies are in italics and boldface. Symbols for species groups are given in figure 2A.
Fig. 4.—Phylogenetic hypothesis of species in the genus Acropora based on the data sets presented in figure 2A and B with major tectonic and climatological events superimposed on it (see text for details)
We thank Jason MacKenzie, Cathie Page, Onno Diekmann, and Wiebe Kooistra for their contributions to the collection of samples. We further thank Charlie Veron for fruitful discussions. This project was funded by the Australian Research Council and James Cook University. M.J.H.v.O. acknowledges receipt of a James Cook postdoctoral fellowship.
References
Adachi J., M. Hasegawa,
Arnason U., R. Spilliaert, A. Palsdottir, A. Arnason,
Audley-Charles M. G.,
Babcock R. C., G. D. Bull, P. L. Harrison, A. J. Heyward, J. K. Oliver, C. C. Wallace, B. L. Willis,
Benzie J. A. H.,
Broughton R. E., T. E. Dowling,
Budd A. F., K. G. Johnson,
Budd A. F., T. A. Stemann, K. G. Johnson,
Callaerts P., G. Halder, W. J. Gehring,
Carbone F., R. Matteucci, J. S. Pignatti, A. Russo,
Catmull J., D. C. Hayward, N. E. McIntyre, J. S. Reece-Hoyes, R. Mastro, P. Callaerts, E. E. Ball, D. J. Miller,
Chappell J., A. Omura, T. Esat, M. McCulloch, J. M. Pandolfi, Y. Ota, B. Pillans,
Coates A. G., J. A. Obando,
Diekmann O. E., R. P. M. Bak, W. T. Stam, J. L. Olsen.,
Dowling T. E., B. D. DeMarais,
Dowling T. E., W. R. Hoeh,
Duque-Caro H.,
Fukami H., M. Omori, M. Hatta,
Fumagalli L., P. Taberlet, L. Favre, J. Hausser,
Grant V.,
Hall R., J. D. Holloway,
Haq B. U., J. Hardenbol, P. R. Vail,
Harrison P. L., R. C. Babcock, G. D. Bull, J. K. Oliver, C. C. Wallace, B. L. Willis,
Hatta M., H. Fukami, W. Wang, M. Mori, K. Shimoike, T. Hayashibara, Y. Ina, T. Sugiyama,
Heyward A. J., R. C. Babcock,
Hillis D. M., J. P. Huelsenbeck,
Hoelzel A. R., J. M. Hancock, G. A. Dover,
Hoelzer G. A., D. J. Melnick,
Holmquist R.,
Hughes T. P., D. J. Ayre, J. H. Connell,
Keigwin L.,
Kenyon J. C.,
Kishino H., M. Hasegawa,
Knowlton N., J. L. Mat, H. M. Guzman, R. Rowan,
Knowlton N., L. A. Weigt, L. Anibal Solorzano, D. K. Mills, E. Bermingham,
Lamb T., J. C. Avise,
McMillan W. O., S. R. Palumbi,
McNeill D. F., A. F. Budd, F. P. Borne,
Miller K., R. C. Babcock,
Miller K. J., J. A. H. Benzie,
Odorico D. M., D. J. Miller,
Oliver J. K., R. C. Babcock,
Palumbi S. R.,
Parani M., C. S. Rao, N. Mathan, C. S. Anuratha, K. K. Narayanan, A. Parida,
Por F. D.,
Rieseberg L. H., D. E. Soltis,
Rögl F., F. F. Steiniger,
Rosen B. R.,
Swofford D. L.,
van Oppen M. J. H., N. R. Hislop, P. J. Hagerman, D. J. Miller,
van Oppen M. J. H., B. L. Willis, D. J. Miller,
van Oppen M. J. H., B. L. Willis, H. W. J. A. van Vugt, D. J. Miller,
Veron J. E. N.,
Veron J. E. N., C. C. Wallace,
von Fritsch J.,
Wallace C. C.,
Wells J. W.,
Willis B. L., R. C. Babcock, P. L. Harrison, J. K. Oliver, C. C. Wallace,