-
PDF
- Split View
-
Views
-
Cite
Cite
Riichiro Yoshida, Tokunori Hobo, Kazuya Ichimura, Tsuyoshi Mizoguchi, Fuminori Takahashi, Jose Aronso, Joseph R. Ecker, Kazuo Shinozaki, ABA-Activated SnRK2 Protein Kinase is Required for Dehydration Stress Signaling in Arabidopsis, Plant and Cell Physiology, Volume 43, Issue 12, 15 December 2002, Pages 1473–1483, https://doi.org/10.1093/pcp/pcf188
Close - Share Icon Share
Abstract
Protein phosphorylation has pivotal roles in ABA and osmotic stress signaling in higher plants. Two protein phosphatase genes, ABI1 and ABI2, are known to regulate these signaling pathways in Arabidopsis. The identity of ABA-activated protein kinases required for the ABA signaling, however, remains to be elucidated. Here we demonstrate that two protein kinases, p44 and p42, were activated by ABA in Arabidopsis T87 cultured cells, and at least one protein kinase, p44, was activated not only by ABA but also by low humidity in Arabidopsis plants. Analysis of T-DNA knockout mutants and biochemical analysis using a specific antibody revealed that the p44 is encoded by a SnRK2-type protein kinase gene, SRK2E. The srk2e mutation resulted in a wilty phenotype mainly due to loss of stomatal closure in response to a rapid humidity decrease. ABA-inducible gene expression of rd22 and rd29B was suppressed in srk2e. These results show that SRK2E plays an important role in ABA signaling in response to water stress.
(Received October 22, 2002; Accepted November 9, 2002)
Introduction
In higher plants, ABA plays a pivotal role in the response to water stress (Bray 1997, Bonetta and McCourt 1998, Shinozaki and Yamaguchi-Shinozaki 1997, Shinozaki and Yamaguchi-Shinozaki 2000, Finkelstein et al. 2002) and several developmental processes, including embryo maturation (Holdsworth et al. 1999) and seed dormancy (Bewley 1997). Various signaling factors participating in ABA signal transduction have been identified and analyzed at the whole plant, tissue and cellular levels: for example, cyclic ADP-ribose (Leckie et al. 1998, Wu et al. 1997), phospholipase D (Ritchie and Gilroy 1998, Ritchie and Gilroy 2000), phospholipase C (Staxen et al. 1999), syntaxin (Leyman et al. 1999), hydrogen peroxide (Pei et al. 2000), inositol phosphatase (Xiong et al. 2001a), GTPase (Lemichez et al. 2001), GTP-binding protein (Wang et al. 2001), mRNA cap binding protein (Hugouvieux et al. 2001) and Sm-like protein (Xiong et al. 2001b). In addition, many genes that respond to ABA have been identified in various plant species and their promotor regions analyzed in detail (Marcotte et al. 1989, Mundy et al. 1990, see reviews Bray 1997). Using the yeast one-hybrid system, the bZip-type transcription factors that specifically bind to the ABA-responsive cis elements (ABREs) were shown to function downstream of ABA signaling for the induction of specific genes (Hobo et al. 1999, Uno et al. 2000, Choi et al. 2000, Finkelstein and Lynch 2000, Lopez-Molina and Chua 2000).
Genetic analysis using a series of Arabidopsis mutants with altered ABA sensitivity have also helped to elucidate the kinds of molecules participating in ABA signaling and how they systematize each other in the pathway (Merlot and Giraudat 1997, Bonetta and McCourt 1998). The ABA-insensitive mutant abi1 has been used in the study of ABA signaling because this mutant shows several impaired responses to ABA both in physiological and gene expression levels (Bray 1997, Shinozaki and Yamaguchi-Shinozaki 1997, Shinozaki and Yamaguchi-Shinozaki 2000, Merlot and Giraudat 1997, Finkelstein et al. 2002). The ABI1 gene encodes protein phosphatase 2C (PP2C) and may essentially function upstream of the signaling pathway (Leung et al. 1994, Meyer et al. 1994). This result suggests that protein phosphorylation is involved in ABA signaling. Other investigators have supported this idea (Armstrong et al. 1995, Hey et al. 1997). Using Arabidopsis guard cell protoplasts, Pei et al. (1997) demonstrated that the general protein kinase inhibitor K252a partially suppressed the abi1 phenotype in loss of ABA-dependent stomatal closure. In potato, ABA-inducible wounding genes are inhibited by staurosporine (Damman et al. 1997). These reports support the possibility of the existence of ABA-activated protein kinase(s) in plant cells. Although several other ABA-responsive mutants have been found, the mutant genes were restricted to PP2C (abi2: Leung et al. 1997), transcription factors (abi3, abi4, abi5: Giraudat et al. 1992, Finkelstein et al. 1998, Finkelstein and Lynch 2000, Lopez-Molina and Chua 2000), and farnesyl transferase (era1: Cutler et al. 1996, Pei et al. 1998); none of the mutants functionally related to the protein phosphorylation have been isolated to date.
Protein kinases that are specifically activated by ABA should help to elucidate the mechanism of ABA signaling in plant cells. Anderberg and Walker-Simmons (1992) isolated PKABA1 from wheat. PKABA1 belongs to SNF1 type of protein kinase and this protein kinase suppresses the induction of α-amylase and GAMyb genes by GA in barley aleurone cells (Gomez-Cadenas et al. 1999). Knetsch et al. (1996) found MAPK activity in ABA-treated barley aleurone protoplast. Mori and Muto (1997) clarified an ABA-activated 48-kDa protein kinase in fava bean guard cells. Using a biochemical approach, Li et al. (2000) succeeded in purifying the 48-kDa ABA-activated protein kinase, and identified the corresponding gene, AAPK (Li and Assmann 1996) in fava bean guard cells. They clearly demonstrated that AAPK did actually regulate ABA-induced activation of plasma membrane anion channels in guard cells and concomitant response to their closure.
Conserved N-terminal regions of AREB1 and AREB2 bZip type ABRE-binding proteins are specifically phosphorylated by 42-kDa ABA-activated protein kinase in Arabidopsis T87 cultured cells (Uno et al. 2000). The ABA-dependent phosphorylation of ABI5 has also been demonstrated (Lopez-Molina et al. 2001). These findings suggest the existence of ABA-dependent protein kinase(s) and their function in Arabidopsis cells. Using histone as an in-gel kinase assay substrate, we also found the 42- and 44-kDa protein kinases that were rapidly and specifically activated by ABA in Arabidopsis T87 cells. We searched for the candidate genes corresponding to our kinases in the Arabidopsis genome now available, and finally focused on SnRK2 (SNF1-related protein kinase 2) genes (Halford et al. 2000). One of the SnRK2 genes, SRK2E, was found to have essential roles in ABA and water stress signaling because its T-DNA knockout mutant, srk2e, revealed a wilty phenotype against rapid humidity decrease. We discuss the roles of SRK2E in ABA-signaling from physiological and biochemical aspects.
Results
ABA specifically activates p42 and p44 protein kinases in T87 cells
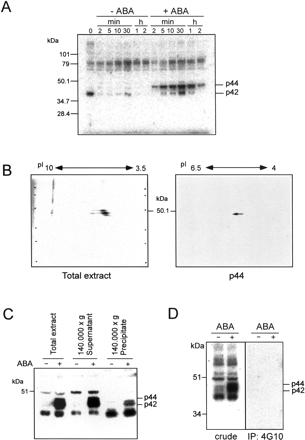
To confirm whether ABA specifically activates protein kinases in Arabidopsis, we performed an in-gel kinase assay using protein extracts prepared from T87 cells treated with 50 µM ABA for indicated times. ABA treatment specifically activated the 42- and 44-kDa protein kinases (p42 and p44) (Fig. 1A). p44 was activated within 2 min, and p42 began to be activated after 5 min of ABA treatment. Protein kinase activities of both p42 and p44 peaked 30 min after ABA treatment.
On-line based search for a candidate gene encoding p44
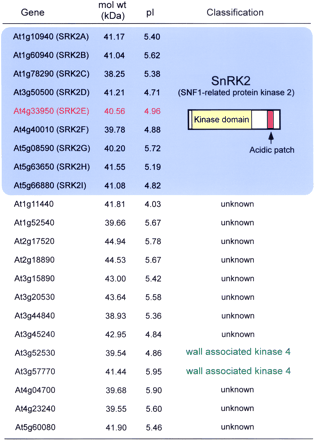
In order to identify ABA-activated protein kinase, we purified p44 and analyzed its sequence. Contrary to our expectation, the sequence data did not contain any protein kinase motifs (data not shown). Thus, we used other methods to identify the protein kinase. To determine the pI value, we performed 2-D PAGE using protein extracts prepared from ABA-treated T87 cells, and then performed an in-gel kinase assay. As shown in Fig. 1B, these kinases had pI values of 4–6. We partially purified p44 protein from ABA-treated cell extract, and subjected it to 2-D PAGE using narrow range Ampholyte (pH 4.0–6.5, Amersham) for IEF. The subsequent in-gel kinase assay revealed that pI of p44 was focused around 5 (Fig. 1B). We also examined whether these kinases are soluble or insoluble proteins. T87 cell protein extracts were ultracentrifuged at 140,000×g, and their supernatant and precipitates were subjected to the in-gel kinase assay. As shown in Fig. 1C, most of the kinase activity remained in the supernatant fraction. These results suggest that two kinases exist in the cytosol, not in the membrane in Arabidopsis cells. Since our ABA-activated protein kinases had a molecular size similar to MAPKs identified in several plant species, we examined whether these kinases are phosphorylated at the Tyr residue. T87 cell extracts were subjected to an immunoprecipitation assay using a phosphotyrosine-specific monoclonal antibody 4G10. Immunoprecipitate complex was subjected to the in-gel kinase assay. As revealed in Fig. 1D, distinct signals were not found in ABA-treated cell extracts after immunoprecipitation. This indicates that, p42 and p44 are not protein kinases whose activation is determined by its Tyr phosphorylation, namely, MAPKs. Based on four biochemical properties of the ABA-activated protein kinases including Ser/Thr kinase, mol wt (42, 44-kDa), pI value (pI 5), and soluble protein, we examined how many genes correspond to those four characteristics in the Arabidopsis genome. We first used the TIGR Arabidopsis database, and searched for kinase genes based on their annotation database. We found 602 kinase genes other than receptor kinase genes (data not shown), and then examined each of their predicted mol wt and pI according to the MIPS database (MAtDB). We selected genes with a mol wt between 38 and 45-kDa, pI between 4 and 6.5, and encoding Ser/Thr type kinase. Finally, 22 candidate genes were picked up (Fig. 2). Among these genes, nine genes were SnRK2 protein kinase (SNF1-related protein kinase 2). We named these nine genes as SRK2 in alphabetical order (SRK2A – SRK2I). Since both PKABA1 (Anderberg and Walker-Simmons 1992) and AAPK (Li et al. 2000) belong to SnRK2 and have been known to function in ABA signaling, we further analyzed these nine genes.
ABA activates SRK2E in Arabidopsis cells and plants
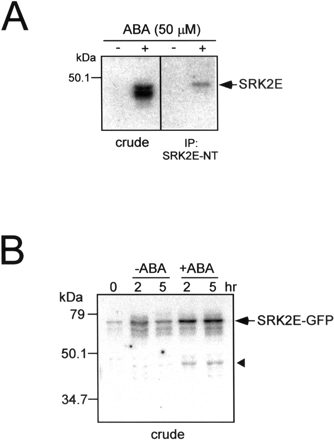
To examine whether the SnRK2 proteins are activated by ABA in Arabidopsis cells, we overexpressed SnRK2 genes in T87 cells as SnRK2–GFP fusion protein and analyzed their ABA-dependent activity. ABA specifically activated SRK2E (At4g33950; shown by red letter in Fig. 2) in T87 cells (data not shown). Mol wt and pI value of SRK2E are 40.56 and 4.96, respectively. We therefore made an antibody for the N-terminal amino acid sequence of SRK2E (SRK2E-NT) to perform an immuno-kinase assay. Protein extracts from T87 cells treated with ABA for 30 min were immunoprecipitated with SRK2E-NT, and its immuno-complex was subjected to an in-gel kinase assay. As shown in Fig. 3A, SRK2E-NT specifically interacted with p44 in ABA-treated T87 cells. To confirm the SRK2E activity in Arabidopsis plants, we constructed transgenic plants that overexpress SRK2E–GFP fusion protein (35S::SRK2E-GFP). The transgenic plants were treated with ABA for 2 and 5 h, and then their leaf protein extracts were subjected to an in-gel kinase assay. Since the predicted mol wt of this fusion protein is around 70 kDa, we could easily separate the fusion protein from endogenous SRK2E to detect its activity by an in-gel kinase assay. We detected the protein kinase activity of SRK2E–GFP with a mol wt of 70 kDa that was activated by ABA application (Fig. 3B). We also detected ABA-dependent activation of protein kinase activity at a mol wt between 40 and 47 kDa. This mol wt corresponds to that of endogenous SRK2E. These results suggest that the p44 kinase activated in ABA-treated T87 cells was SRK2E.
ABA-dependent stomatal closure is impaired in srk2e
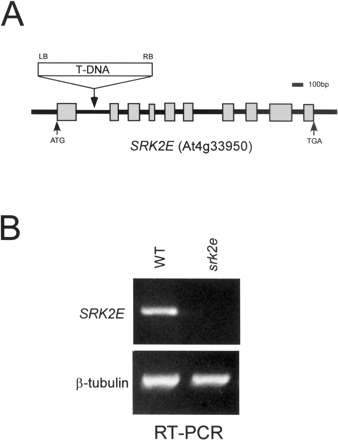
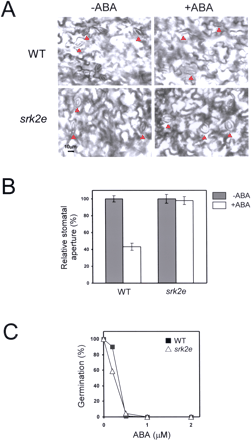
We performed loss-of-function analysis of SRK2E by using T-DNA knockout mutant, srk2e. RT-PCR analysis clearly demonstrated that SRK2E gene was not expressed in the srk2e mutant (Fig. 4). We examined whether ABA normally induced stomatal closure in srk2e. While ABA did actually close the stomata in the wild type, all the stomata in srk2e remained open (Fig. 5A, B). We next examined the inhibitory effect of ABA on seed germination in srk2e. However, srk2e had almost the same levels of sensitivity to ABA as the wild type (Fig. 5C). These results indicate that SRK2E specifically functions in ABA-dependent stomata closure at the seedling stage, but not in the germination stage.
srk2e revealed wilty phenotype under rapid humidity decrease
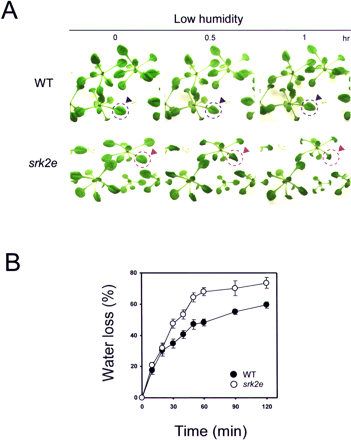
Since the srk2e mutant showed loss of ABA-dependent stomatal closure, we examined the effect of water stress in srk2e plants. Wild-type and mutant seedlings growing on agar in Petri dishes were exposed to low-humidity stress by opening the lid of the Petri dishes. Within 30 min of the treatment, srk2e leaves began to shrink, and then wilted drastically after 1 h (Fig. 6A). To assess water loss from seedlings, we detached the aerial part of 2-week-old seedlings and measured the fresh weight. The srk2e mutant clearly showed the severe loss of fresh weight as compared to the wild type (Fig. 6B). This result strongly indicated that, in srk2e, water was more easily and rapidly lost in response to a decrease in humidity. A relatively higher rate of water loss was confirmed in wild-type seedlings within 20 min of stress treatment, and then the degree gradually lowered. In srk2e, a large rate of water loss continued after 30–60 min of stress. These results indicated that the wilty phenotype in srk2e is due mainly to loss of stomatal closure in spite of a rapid osmotic change.
The SRK2E protein is activated by low humidity
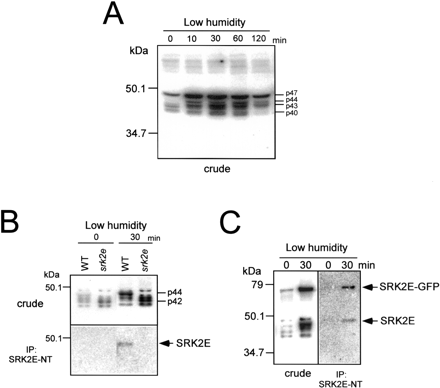
To determine how SRK2E contributes to water stress signaling in guard cell closure, we examined whether this protein is activated by low humidity. We examined the changes in protein kinase activities in wild-type leaf tissue against low-humidity stress. The in-gel kinase assay revealed that at least four kinases (p47, p44, p43, p40) were rapidly and transiently activated by humidity change in wild-type seedlings (Fig. 7A). We then analyzed srk2e mutant seedlings to see how activation profiles are changed under the same humidity stress. As shown in Fig. 7B, p44 activity was not detected in srk2e treated with low humidity. Activities of protein kinases with molecular mass around 42 kDa increased in srk2e under stress conditions. Next, we used SRK2E-NT to perform an immuno-kinase assay. Protein extracts from leaves treated with low-humidity stress for 30 min were used for immunoprecipitation coupled with in-gel kinase assay. Autoradiography clearly indicated that the SRK2E-NT specifically interacted with p44 in wild-type plant extract but not in the srk2e extract (Fig. 7B). We then treated low-humidity stress to 35S::SRK2E-GFP plants. Fig. 7C clearly shows that SRK2E–GFP fusion protein was actually activated by 30 min of low-humidity stress. SRK2E-NT also recognized this activated SRK2E–GFP as shown by immunoprecipitation assay (Fig. 7C). These results indicated that SRK2E was activated not only by ABA but also by low-humidity stress.
Induction of rd29B and rd22 genes by ABA was blocked in srk2e
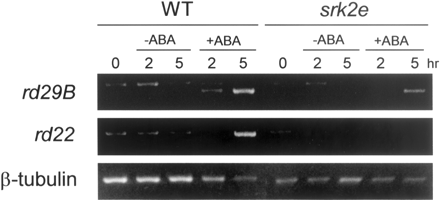
In this study, we found that SRK2E had critical roles in ABA signaling, especially in the guard cell response of seedlings, but not in seed germination. To examine the ABA-dependent gene expression in srk2e seedlings, we used two ABA-inducible genes, rd29B and rd22 (Yamaguchi-Shinozaki and Shinozaki 1993a, Yamaguchi-Shinozaki and Shinozaki 1993b). Total RNAs were prepared from ABA-treated srk2e and wild-type seedlings, and then subjected to RT-PCR analysis to detect rd29B and rd22 transcripts. We clearly detected ABA-dependent expression of both genes in wild-type seedlings (Fig. 8). In contrast, ABA-inducible gene expression of rd29B and rd22 was blocked in the srk2e mutant. We failed to detect the rd22 transcript in ABA-treated srk2e seedlings. This result indicates that SRK2E is required to control induction of ABA-responsive genes.
Discussion
Protein phosphatases have been shown to function in ABA signal transduction using abi1 and abi2 mutants (Leung et al. 1994, Leung et al. 1997, Meyer et al. 1994, Pei et al. 1997). However, no protein kinase has been identified that functions in ABA signaling and is activated by ABA in Arabidopsis. We have been interested in protein phosphorylation in ABA signaling, and in identifying ABA-dependent protein kinase activities in Arabidopsis. To identify the kinase genes in Arabidopsis T87 cultured cells, we examined several biochemical features of the p44 protein kinase: mol wt (44 kDa), pI 5, soluble protein, and Ser/Thr kinase (Fig. 1). Using the Arabidopsis genome database, we selected 22 candidate genes meeting our defined conditions (Fig. 2). Among them, we found nine SnRK2 genes. Several reports have indicated the importance of SnRK2 in ABA or osmotic signaling in wheat (Gomez-Cadenas et al. 1999), fava bean (Li et al. 2000) and tobacco (Mikolajczyk et al. 2000). In contrast, no protein kinases that function in ABA signaling have been identified in Arabidopsis. We have found that one of the SnRK2, SRK2E (At4g33950), was specifically activated by ABA in Arabidopsis cells or plants (Fig. 3). SRK2E knockout mutant srk2e (Fig. 4) lost the ABA-dependent stomatal closure and also showed a wilty phenotype under low humidity stress (Fig. 5, 6). Our data suggested that the wilty phenotype observed in srk2e is due to repressed stomatal closure in low-humidity conditions. Significant water loss of srk2e occurred after 30 min of low-humidity stress (Fig. 6B). The peak of transient activation of p44 activity in leaf tissues was observed after 30 min of low-humidity stress (Fig. 7A). The timing of the activation of p44 coincided with that of severe water loss of srk2e. We clearly showed that the 44-kDa kinase activity (p44) is specifically lost in low-humidity stressed srk2e leaves. We also identified p44 as SRK2E by immuno-complex kinase activity assay (Fig. 3A, 7B, 7C). These results suggest that SRK2E functions in ABA signaling of stomatal closure. We confirmed that the wilty phenotype in srk2e was not so severe as compared to abi1-1 and abi2-1 mutants (data not shown). As shown in Fig. 7A, at least four kinases were activated by a rapid humidity change. These protein kinases may redundantly function in dehydration stress signaling, and this redundancy may be considered to generate the weak phenotype in srk2e. We are also planning to examine the functional relationships between SRK2E and ABI1 or SRK2E and ABI2.
SRK2E has sequence similarity with fava bean AAPK that functions in ABA signaling in guard cells (Li et al. 2000). Expression of AAPK was specific to fava bean guard cells, whereas we detected SRK2E transcripts in leaves, roots and seeds by RT-PCR (data not shown). We analyzed the expression of two ABA-inducible genes, rd29B and rd22, in srk2e seedlings. ABA-inducible expression of these two genes was significantly suppressed in srk2e (Fig. 8). This suggests that SRK2E functions in ABA signaling upstream of ABA-responsive gene expression of rd29B and rd22 as well as in stomata closure. We observed ABA-responsive phosphorylation of AREB1 and AREB2 (ABA Responsive Element Binding Proteins 1, 2) which encode bZip transcription factors involved in ABA-dependent expression of rd29B (Uno et al. 2000). Microarray analysis will give us more information on the role of SRK2E in ABA-dependent gene expression.
Recently, Li et al. (2002) identified an AAPK-interacting protein, AKIP1, that contains an RNA-binding domain. They found that AKIP1 was specifically phosphorylated by ABA-activated AAPK to increase the affinity of AKIP1 for dehydrin transcript. They proposed an important role of AAPK in post-transcriptional RNA metabolism in guard cells. In contrast, SRK2E functions in transcriptional regulation of ABA-inducible genes in seedlings, which suggests that SnRK2 protein kinases are involved in several processes of ABA signal transduction including transcriptional as well as post-transcriptional regulation.
We detected several protein kinases with a molecular mass of 40–47 kDa that are activated by low humidity in Arabidopsis seedlings (Fig. 7A). In srk2e mutant, p44 protein kinase (SRK2E) activity was lost whereas p42 protein kinase activity was rather enhanced, which suggests existence of several redundant protein kinases in ABA signal transduction pathways. Among them, AtGSK1, an Arabidopsis homolog of GSK3/shaggy-like kinase, functions in high-salinity stress tolerance (Piao et al. 2001). Recently, Guo et al. (2002) showed that PKS3 (protein kinase S3), an Arabidopsis homolog of SOS2 protein kinase, functions as a global negative regulator of ABA signaling. PKS3 physically interacts with ABI1 and ABI2 protein phosphatases. It is also possible that other protein kinases we have selected in Fig. 2, including SnRK2, are activated by ABA. These different protein kinases may be involved in ABA signal transduction pathways, which make the ABA response very complex.
SRK2E specifically functions at the seedling stage, not at the germination stage (Fig. 5C). ABI1 and ABI2 function at both germination and seedling stages, whereas ABI3 and ABI5 function only at the germination stage. Different signaling components may function in different cells or tissues in ABA signal transduction. These results also provoke the complexity in ABA-signaling as mentioned elsewhere (Bonetta and McCourt 1998, Finkelstein et al. 2002).
To elucidate the function of SRK2E in ABA signal transduction in seedling stage, we are analyzing genetic as well as protein–protein interactions between SRK2E and several already known proteins involved in ABA signaling, such as ABI, and ERA1. We will use double mutants and a two-hybrid system to examine further the role of SRK2E in ABA signaling.
Materials and Methods
Plant growth conditions
The Columbia ecotype of Arabidopsis thaliana was used. The seeds of wild-type (Columbia ecotype), srk2e and transgenic plants were sterilized with 70% ethanol and 1% Antiformin and sown on GM agar plates (Valvekens et al. 1988) containing no sucrose. They were grown under 16 h light/8 h dark conditions at 22°C for 3 weeks.
Generation of transgenic plants
The SRK2E–GFP transgenic plants were generated by fusing the SRK2E coding region in frame to GFP driven by CaMV 35S promotor in the pBE2113 vector (Mitsuhara et al. 1996). The transformation vector was introduced into Agrobacterium, and transformed into wild-type Arabidopsis (Columbia ecotype) plants by floral infiltration.
Water loss measurement
Rosette leaves of wild-type and srk2e seedlings were detached from their roots and placed on weighing dishes (Tan et al. 1997). These dishes were left on the lab bench, and then loss in fresh weight was monitored at the indicated times.
Stomatal aperture measurements
Leaves of wild-type and srk2e seedlings were detached and pre-incubated for 1 h in a buffer containing 10 mM MES-Tris (pH 6.1), 30 mM KCl under light condition. Then, these leaves were moved to a solution containing 50 µM ABA (mixed isomers; Wako) in the same buffer and then incubated for 1 h. Guard cells were observed using a Axioplan 2 microscope (Carl Zeiss). We measured the width and length of each stomatal pore in the picture.
ABA sensitivity at seed germination
Seeds were sterilized as mentioned above, and sown on an MS agar plate containing various concentrations of ABA. These seeds were chilled for 4 d at 4°C in darkness prior to germination (Gosti et al. 1999).
Low humidity treatment
We removed the lid of an agar plate to induce low-humidity stress. This experiment was performed at the same place on the lab bench. Air humidity was always between 45 and 50% at the stress period.
ABA treatment in Arabidopsis T87 cells and plants
T87 cells (10 ml) on the 10th day in culture were transferred into a 15-ml plastic tube and ABA solution was added to the final concentration of 50 µM. At indicated time points, these cells were harvested by filtration. The cells were quickly frozen in liquid nitrogen and stored at –80°C. For plants, 3-week-old seedlings were carefully pulled out from the GM agar plates and then their roots were soaked in distilled water in FALCON 351007 Petri dishes. These seedlings were left for 24 h to extrude mechanical stress. After this operation, water was carefully removed by pipetting and then, 5 ml of ABA solution were gently added to the Petri dishes so that the roots are completely soaked in this solution. After designated time points, the seedlings were quickly picked up from the solution and put onto a paper towel to remove excess solution on the roots. These seedlings were put into 1.5-ml tubes, quickly frozen in liquid nitrogen and stored at –80°C.
Protein extraction
Protein extraction was performed as described previously (Ichimura et al. 2000).
Two-dimensional electrophoresis
Protein samples were suspended in lysis buffer containing 9.5 M urea, 2% Triton X-100, 2% Ampholines (pH 3.5–10 and pH 4–6). These samples were subjected to first-dimensional separation by isoelectric focusing in rod gels. After equilibration, these rod gels were loaded onto a second-dimensional SDS-PAGE gel.
In-gel kinase assay
The in-gel kinase assay was performed as described previously (Zhang and Klessig 1997, Ichimura et al. 2000). The concentration of the SDS-PAGE gel was 10%. Histone was used as a kinase substrate.
Antibody production
SRK2E-specific antibody (SRK2E-NT) was produced against synthetic peptides corresponding to the N-terminus of SRK2E (MDRPAVSGPMDLC). Polyclonal antisera were raised in rabbits and purified by affinity chromatography (Peptide Institute Inc, Osaka, Japan).
Immunoprecipitation
Immunoprecipitation was performed as described previously (Ichimura et al. 2000). Leaf crude extract (500 µg) was used in this assay.
RNA extraction and RT-PCR analysis
Total RNA was extracted from rosette leaves by using commercial kit (Total RNA Extraction Kit; Amersham). RT-PCR was performed by commercial kit (mRNA Selective RT-PCR Kit; TAKARA). Following primers were used in this analysis: SRK2E [5′-ATGGATCGACCAGCAGTGAG-3′ for forward; 5′-CATTGCGTACACAATCTCTC-3′ for reverse], rd29B [5′-AATTATCAGTCCAAAGTTACTGAT-3′ for forward; 5′-TTTCTGCCCGTAAGCAGTAACAGA-3′ for reverse] and rd22 [5′-ATGGCGATTCGGCTTCCTCT-3′ for forward; 5′-GTAGCTGAACCACACAACAT-3′ for reverse].
Acknowledgments
We thank Dr. Tokihiko Nanjo for advice and helpful discussions. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences (BRAIN). This work was supported in part by a grant for Genome Research from RIKEN, the Program for Promotion of Basic Research Activities for Innovative Biosciences, the Special Coordination Fund of the Science and Technology Agency to K.S. It was also supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (No. 14740451 to R.Y).
Present address: The Sainsbury Laboratory, John Innes Center, Norwich NR4 7UH, U.K.
Present address: Institute of Biological Sciences, Tsukuba University, Tennodai, Tsukuba, Ibaraki, 305-8572, Japan
Corresponding author: E-mail, sinozaki@rtc.riken.go.jp; Fax, +81-298-36-9060.
Fig. 1 ABA activates protein kinases in Arabidopsis T87 cells. (A) T87 cells were treated with or without 50 µM ABA for indicated times and their protein extracts were prepared. The kinase activities were determined by an in-gel kinase assay using histone as a substrate. Total cell extracts (10 µg) were loaded in each lane. (B) Protein extracts from ABA-treated T87 cells were applied to 2-D PAGE and subsequently subjected to in-gel kinase assay. Total protein (left) and partially purified p44 protein (right) were analyzed. (C) T87 cell extracts were ultracentrifuged at 140,000×g for 1 h, and then their supernatants and precipitates were subjected to an in-gel kinase assay. Proteins (10 µg) were assayed in each fraction. (D) Total cell extracts (500 µg) were immunoprecipitated with 10 µg of the phospho-Tyr-specific monoclonal antibody 4G10. The immune complex was subsequently subjected to an in-gel kinase assay with histone as a substrate.
Fig. 2Arabidopsis Ser/Thr protein kinases whose mol wt (38–45 kDa) and pl (4–6.5) corresponded to p44 kinase.
Fig. 3 SRK2E is activated by ABA in both Arabidopsis cells and plants. (A) T87 cells were treated with 50 µM ABA for 30 min. Protein extracts (300 µg) were immunoprecipitated with SRK2E-specific antibody SRK2E-NT. Kinase activity of the crude extracts (10 µg) and immune complex were subsequently analyzed by an in-gel kinase assay with histone as a substrate. (B) 35S::SRK2E-GFP plants were treated with 50 µM ABA for indicated times as described in Materials and Methods. Leaf extracts (20 µg) were subjected to an in-gel kinase assay with histone as a substrate. Triangle indicates protein kinase corresponding to endogenous SRK2E.
Fig. 4 T-DNA knockout mutant of srk2e. (A) Map of SRK2E gene (At4g33950). T-DNA is inserted in the first intron. (B) RT-PCR analysis of SRK2E in wild type (WT) and srk2e. Total RNAs (1 µg) extracted from 3-week-old seedlings were used.
Fig. 5 SRK2E affects ABA-signaling in stomatal closure, but not in germination stage. (A) Effect of ABA (50 µg) on stomatal closure in wild type (WT) and srk2e. ABA was treated for 1 h. Stomata cells were indicated at red arrows. (B) Relative stomatal aperture in response to 50 µM ABA for 1 h. Data represent the mean ± SE, 40–60 stomata were measured in each process. (C) ABA dose response for germination inhibition. Seeds of wild-type and srk2e were plated on agar plates supplemented with the indicated concentration of ABA. These seeds were incubated for 3 d at 22°C in 16 h light/8 h dark conditions. Greened cotyledons were judged as germinated seeds. The number of germinated seeds was represented as the percentage of the total number of seeds tested (50–80). Essentially same results were obtained in more than two independent experiments.
Fig. 6 srk2e mutant shows wilty phenotype under low humidity stress condition. (A) Wild type (WT: upper) and srk2e (lower) seedlings under low-humidity stress. As indicated by dotted circles (blue: WT, red: srk2e), srk2e leaves are susceptible to low humidity. (B) Water loss in WT and srk2e rosette leaves (mean ± SE, n = 5).
Fig. 7 SRK2E is activated by low-humidity stress. (A) Wild type (WT) seedlings were treated with low humidity for indicated times. Total protein extracts (20 µg) prepared from their leaves were applied to the in-gel kinase assay with histone as a substrate. p47, p44, p43 and p40 indicate four protein kinases which are activated by low humidity. (B) WT and srk2e plants were treated with low humidity for 30 min and their leaf extracts were prepared. Leaf extracts (500 µg) were immunoprecipitated with SRK2E-specific antibody SRK2E-NT. Kinase activity of the crude extracts (20 µg) and immune complex were subsequently analyzed by an in-gel kinase assay with histone as a substrate. p44 shows a protein kinase whose activity is not detected in srk2e. p42 indicates a protein kinase whose activity is increased in srk2e. (C) 35S::SRK2E-GFP plants were treated with low humidity and their leaf extracts were prepared. Leaf extracts (500 µg) were immunoprecipitated with SRK2E-specific antibody SRK2E-NT. Kinase activity of the crude extracts (20 µg) and immune complex were subsequently analyzed by an in-gel kinase assay with histone as a substrate. Kinase activity shown at high mol wt (around 70 kDa) is SRK2E–GFP.
Fig. 8 ABA-inducible gene expressions were suppressed in srk2e. Wild type (WT) and srk2e seedlings were treated with or without 50 µM ABA for indicated times as described in Materials and Methods, and their total RNAs were extracted. Total RNAs (1 µg) were used for RT-PCR reaction to confirm the expression of rd29B and rd22. Primers for β-tubulin were used in the PCR reactions as internal controls.
Abbreviations
References
Anderberg, R.J. and Walker-Simmons, M.K. (
Armstrong, F., Leung, J., Grabov, A., Brearley, J., Giraudat, J. and Blatt, M.R. (
Bonetta, D. and McCourt, P. (
Choi, H., Hong, J., Ha, J., Kang, J. and Kim, S.Y. (
Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S. and McCourt, P. (
Damman, C., Rojo, E. and Sanchez-Serrano, J.J. (
Finkelstein, R.R., Gampala, S.S.L. and Rock, C.D. (
Finkelstein, R.R. and Lynch, T.J. (
Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S. and Goodman, H.M. (
Giraudat, J., Hauge, B., Valon, C., Smalle, J., Parcy, F. and Goodman, H. (
Gomez-Cadenas, A., Verhey, S.D., Holappa, L.D., Shen, Q., Ho, T.H. and Walker-Simmons, M.K. (
Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A.R., Vartanian, N. and Giraudat, J. (
Guo, Y., Halfter, U., Ishitani, M. and Zhu, J.K. (
Guo, Y., Xiong, L., Song, C.P., Gong, D., Halfer, U. and Zhu, J.K. (
Halford, N.G., Bouly, J.P. and Thomas, M. (
Hey, S.J., Bacon, A., Burnett, E. and Neill, S.J. (
Hobo, T., Kowyama, Y. and Hattori, T. (
Holdsworth, M., Kurup, S. and McKibbin, R. (
Hugouvieux, V., Kwak, J.M. and Schroeder, J.I. (
Ichimura, K., Mizoguchi, T., Yoshida, R., Yuasa, T. and Shinozaki, K. (
Knetsch, M.L.W., Wang, M., Snaar-Jagalska, B.E. and Heimovaara-Dijkstra, S. (
Leckie, C.P., McAinsh, M.R., Allen, G.J., Sanders, D. and Hetherington, A.M. (
Lemichez, E., Wu, Y., Sanchez, J.P., Mettouchi, A., Mathur, J. and Chua, N.H. (
Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F. and Giraudat, J. (
Leung, J., Merlot, S. and Giraudat, J. (
Leyman, B., Geelen, D., Quintero, F.J. and Blatt, M.R. (
Li, J. and Assmann, S.M. (
Li, J., Kinoshita, T., Pandey, S., Ng, C.K.Y, Gygi, S.P., Shimazaki, K. and Assmann, S.M. (
Li, J., Wang, X.Q., Watson, M.B. and Assmann, S.M. (
Lopez-Molina, L. and Chua, N.H. (
Lopez-Molina, L., Mongrand, S. and Chua, N.H. (
Marcotte, W.R., Jr., Russell, S.H. and Quatrano, R.S. (
Meyer, K., Leube, M.P. and Grill, E. (
Merlot, S. and Giraudat, J. (
Mikolajczyk, M., Awotunde, O.S., Muszynska, G., Klessig, D.F. and Dobrowolska, G. (
Mitsuhara, I., Ugaki, M., Hirochika, H., Ohshima, M., Murakami, T., Gotoh, Y., Katayose, Y., Nakamura, S., Honkura, R., Nishimiya, S., Ueno, K., Mochizuki, A., Tanimoto, H., Tsugawa, H., Otsuki, Y. and Ohashi, Y. (
Mori, I.C. and Muto, S. (
Mundy, J., Yamaguchi-Shinozaki, K. and Chua, N.H. (
Pei, Z.M., Ghassemian, Kwak, C.M., McCourt, P. and Schroeder, J.I. (
Pei, Z.M., Kuchitsu, K., Ward, J.M., Schwarz, M. and Schroeder, J. (
Pei, Z.M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G.J., Grill, E. and Schroeder, J.I. (
Piao, H.L., Lim, J.H., Kim, S.J., Cheong, G.W. and Hwang, I. (
Ritchie, S. and Gilroy, S. (
Ritchie, S. and Gilroy, S. (
Shinozaki, K. and Yamaguchi-Shinozaki, K. (
Shinozaki, K. and Yamaguchi-Shinozaki, K. (
Staxen, I., Pical, C., Montgomery, L.T., Gray, J.E., Hetherington, A.M. and McAinsh, M.R. (
Tan, B.C., Schwartz, S.H., Zeevaat, J.A.D. and McCarty, D.R. (
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K. and Yamaguchi-Shinozaki, K. (
Valvekens, D., Van Montagu, M. and Van Lijsebettens, M. (
Wang, X.Q., Ullah, H., Jones, A.M. and Assmann, S.M. (
Wu, Y., Kuzma, J., Marechal, E., Graeff, R., Lee, H.C., Foster, R. and Chua, N.H. (
Xiong, L., Gong, Z., Rock, C.D., Subramanian, S., Guo, Y., Xu, W., Galbraith, D. and Zhu, J.K. (
Xiong, L., Lee, B., Ishitani, M., Lee, H., Zhang, C. and Zhu, J.K. (
Yamaguchi-Shinozaki, K. and Shinozaki, K. (
Yamaguchi-Shinozaki, K. and Shinozaki, K. (