-
PDF
- Split View
-
Views
-
Cite
Cite
Rajeev Vibhakar, Greg Foltz, Jae-geun Yoon, Lorie Field, Hwahyung Lee, Gi-yung Ryu, Jessica Pierson, Beverly Davidson, Anup Madan, Dickkopf-1 is an epigenetically silenced candidate tumor suppressor gene in medulloblastoma, Neuro-Oncology, Volume 9, Issue 2, April 2007, Pages 135–144, https://doi.org/10.1215/15228517-2006-038
Close - Share Icon Share
Abstract
Medulloblastoma is a heterogeneous pediatric brain tumor with significant therapy-related morbidity, its five-year survival rates ranging from 30% to 70%. Improvement in diagnosis and therapy requires better understanding of medulloblastoma pathology. We used whole-genome microarray analysis to identify putative tumor suppressor genes silenced by epigenetic mechanisms in medulloblastoma. This analysis yielded 714 up-regulated genes in immortalized medulloblastoma cell line D283 on treatment with histone deacetylase (HDAC) inhibitor trichostatin A (TSA). Dickkopf-1 (DKK1), a Wnt antagonist, was found to be up-regulated on HDAC inhibition. We examined DKK1 expression in primary medulloblastoma cells and patient samples by reverse transcriptase PCR and found it to be significantly down-regulated relative to normal cerebellum. Transfection of a DKK1 gene construct into D283 cell lines suppressed medulloblastoma tumor growth in colony focus assays by 60% (P < 0.001). In addition, adenoviral vector-mediated expression of DKK1 in medulloblastoma cells increased apoptosis fourfold (P < 0.001). These data reveal that inappropriate histone modifications might deregulate DKK1 expression in medulloblastoma tumorigenesis and block its tumor-suppressive activity.
Medulloblastoma accounts for approximately 20% of all primary pediatric brain tumors (Packer et al., 1999). Although advances in treatment with surgery, radiation, and chemotherapy have increased the five-year survival rate to approximately 70% (Packer et al., 1999, 2003), children younger than three years of age show significantly worse outcomes (Rood et al., 2004). Current medulloblastoma treatments have devastating morbidity associated with them because they lack specificity (Mulhern et al., 2005; Packer et al., 1999); thus, new approaches are needed. Understanding the molecular basis of medulloblastoma pathogenesis may identify signaling pathways for targeted therapy. Recent advances have identified several genetic mechanisms, such as mutations and loss of heterozygosity, leading to tumor suppressor loss in medulloblastoma (MacDonald et al., 2003). However, other mechanisms of tumor suppressor loss have not been extensively studied in medulloblastoma (Lindsey et al., 2005).
Over the past several years, there has been an increasing realization that many tumor suppressor genes are silenced by epigenetic rather than genetic mechanisms (Jones, 2003; Jones and Baylin, 2002). Disruption of epigenetic mechanisms is considered to be closely linked to aberrant expression of cancer-associated genes (Feinberg, 2004; Jones and Laird, 1999). Two fundamental epigenetic changes are associated with transcriptional repression of genes in cancer. These are histone modifications (acetylation, methylation, and phosphorylation) and hypermethylation of CpG motifs in DNA promoter regions (Jones and Baylin, 2002). Abundant evidence supports a closed interplay between DNA methylation and histone modifications for establishing gene silencing (Feinberg, 2004; Laird, 2005). Several recent reports indicate that changes in histone tail modifications can overcome the repressive barrier of DNA methylation (Bachman et al., 2003). This has led to the hypothesis that changes in chromatin remodeling proteins are the primary event in creating a “closed” local chromatin structure associated with repressed transcriptional activity of genes. While there are several reports of DNA methylation in medulloblastoma (Fruhwald et al., 2001; Lindsey et al., 2004), the role of histone modifications in regulating gene expression in medulloblastoma has not previously been described. An extensive characterization of genes silenced due to pathological changes in chromatin structure in medulloblastoma could offer a better chance to develop curative measures.
In the present study, we sought to identify genes activated through pharmacological reversal of histone deacetylation by trichostatin A (TSA)3 in medulloblastoma cells using whole-genome microarray analysis. TSA is a potent histone deacetylase (HDAC) inhibitor. We identified Dickkopf-1 (DKK1) as significantly up-regulated on HDAC inhibition. We confirmed transcriptional silencing of DKK1 in the D283 cell line and, more important, in patient-derived primary medulloblastoma cells, as well as in a panel of tumor tissues. Histone acetylation in the promoter region of DKK1 increased fivefold in response to HDAC inhibition. Reexpressing DKK1 in medulloblastoma cells induced apoptosis and inhibited clonogenic growth, supporting its role in the control of cell growth. These data demonstrate the importance of histone acetylation in regulating gene expression in medulloblastoma, and implicate the dysregulation of DKK1 as a potential component of medulloblastoma pathogenesis.
Abbreviations used are as follows: Ad, adenovirus; ChIP, chromatin immunoprecipitation; DKK1, Dickkopf-1; DMSO, dimethylsulfoxide; GFP, green fluorescent protein; HBSS, Hank's balanced salt solution; HDAC, histone deacetylase; neo, neomycin; qPCR, quantitative PCR; RT-PCR, reverse transcriptase PCR; TSA, trichostatin A.
Materials and Methods
Cells, Tissues, and Culture
D283 medulloblastoma cells were obtained from American Type Culture Collection (Rockville, Md.) and cultured in modified Eagle's medium (Gibco, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (Gibco) according to the supplier's recommendations. Primary cell cultures were derived from biopsy specimens of medulloblastoma patients under a protocol approved by the institutional review board at the University of Iowa Hospitals and Clinics. To generate primary cell cultures, approximately 200-250 mg of tumor tissue was immersed and incubated in 0.05 mM EDTA solution containing 0.05% trypsin (Sigma, St. Louis, Mo.) at 4°C for 8 h. The tissue samples were minced into 0.3 mm3 fragments and suspended in Hank's balanced salt solution (HBSS) containing 4 mg DNase I, 40 mg collagenase IV, and 100 units of hyaluronidase type V (all from Sigma). Single-cell suspensions were then passed through no. 100 nylon mesh, washed twice in HBSS, and added to fibronectin-coated tissue culture flasks. Cultures were maintained at low passage numbers (p2-p4) in modified Eagle's medium supplemented with 10% fetal bovine serum as described above. Normal human cerebellum and medulloblastoma patient samples were obtained from the Pediatric Co-operative Human Tissue Network (Columbus, Ohio). All normal cerebellar samples were from nonmalignant adult brain. All medulloblastoma samples were from pediatric patients (<18 years of age). For detailed data on the normal samples, primary cultures, and patient samples, see supplementary data, Table 1S, in the online version of this article at http://neuro-oncology.dukejournals.org.
Microarray Analysis
The D283 cell line was cultured with either 0.2 μM TSA or dimethylsulfoxide (DMSO) for 9 h to generate gene expression profiles in response to TSA. Total RNA was extracted from treated cells using Trizol (Invitrogen, Carlsbad, Calif.). RNA was further purified using the RNeasy kit (Qiagen, Valencia, Calif.) per the manufacturer's protocol, and purity of RNA was determined by the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, Calif.). Two micrograms of total RNA was reverse-transcribed with the Chemiluminescent RT-IVT Labeling Kit (Applied Biosystems, Foster City, Calif.) and hybridized to a 60-mer whole-genome oligonucleotide microarray (Applied Biosystems) containing 33,202 probes representing 29,098 genes, per the manufacturer's protocol. A total of three microarray hybridizations, one for each biological replicate, were performed per treatment. Data were quantile normalized, and a t-test was applied to data for each gene for statistical significance. Differential gene expression was quantified using the Storey q value method (Storey and Tibshirani, 2003). Spotfire software was used for data visualization, and a cut-off of twofold threshold with a false discovery rate of 1% was used to identify epigenetically regulated genes (Spotfire, Somerville, Mass.). Assay on Demand gene expression reagents (Applied Biosystems) for nine randomly selected genes were used to validate microarray data. Data were submitted to the National Center for Biotechnology Information gene expression omnibus database (www.ncbi.nlm.nih.gov/geo).
Real-Time Quantitative Reverse Transcriptase PCR
RNA was isolated from cells and tissues with Triazol (Invitrogen). Real-time PCR was performed on the ABI PRISM 7900 HT detection system using Taq man reagents (Applied Biosystems) per the manufacturer's recommendations. Gene expression was determined with Assay on Demand gene expression reagents. All assays were done in triplicate.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) analysis was done using primary antibodies to acetylated histone 3 (Upstate Biotechnology, Lake Placid, N.Y.). Control (DMSO) or TSA-treated D283 cells (1 × 106) were incubated with 1% formaldehyde for 10 min to cross-link histones to DNA. Cells were washed with cold PBS, resuspended in lysis buffer (Upstate Biotechnology), and sonicated for 10 sec with continuous output using a Branson sonifier (Branson Ultrasonics, Danbury, Conn.). The lysate was centrifuged for 10 min at 13,200 rpm at 4°C, after which the supernatant was incubated with protein A agarose beads (Upstate Biotechnology) for 2 h. The slurry was removed by centrifugation at 1000 rpm for 1 min. The supernatant was collected and incubated at 4°C overnight in four parts (input control, anti-K9 acetylated histone H3, normal rabbit IgG, or no antibody). The immunoprecipitated complexes were collected and washed, and the cross-links were reversed. The samples were then treated with proteinase K overnight, and DNA was extracted by the phenol chloroform method, ethanol precipitated, and resuspended in 50 μl water. PCR was performed on extracted DNA using primers designed to amplify a 250-bp promoter region. To ensure that PCR amplification was in linear range, each reaction was set up at different dilutions of DNA for varying amplification cycle numbers, and final PCR conditions were selected accordingly. The PCR mixture contained 20 pM of each primer, 1 μl extracted DNA, 0.5 units of Taq DNA polymerase (Eppendorf, Pittsburg, Penn.), 0.2 mM of each deoxyribonucleotide, and 2 mM MgSO4 in a final volume of 50 μl. The PCR was performed with the following cycling parameters: an activation step of 94°C for 3 min, followed by 30 cycles of 94°C for 2 min, 50°C for 2 min, and 68°C for 3 min, with a final extension step of 68°C for 10 min. The promoter region of DKK1 was amplified, and the PCR products were quantified by densitometry and plotted as a ratio of acetylated histone (TSA treatment) to unacetylated histone (DMSO treatment). The assays were done in triplicate.
Construction of Expression Vectors
Full-length open reading frame for DKK1 was PCR amplified from a Mammalian Gene Collection clone (MGC:868, BC001539) and subcloned into the pcDNA3.1D/V5-His-TOPO vector (Invitrogen), and the sequence was verified. The PCR product was also cloned into pAD-5CMVIRESeGFPpA, and its sequence was verified. The clone was recombined in HEK293 cells with pacAD5 9.2-100 to produce recombinant adenovirus (Ad) particles (University of Iowa Gene Transfer Vector Core).
Transfection and Colony Formation Assays
Colony formation assays were performed on soft agar. Cells were plated at 1.5 × 105 per well using six-well plates and transfected with pcDNA3.1D/V5-His-TOPO/DKK1, pcDNA3.1D/V5-His-TOPO/lacZ, or pcDNA3.1D/V5-His-TOPO with no insert (mock control) using Trans It-Neural transfection reagents (Mirus Bio Corp., Madison, Wisc.). At 24 h posttransfection, the cells were selected in media supplemented with G418 (1 mg/ml) and simultaneously harvested to confirm their expression at the mRNA level by real-time PCR. G418-resistant cells were maintained for two weeks in culture. Cells were resuspended in media containing 0.3% agarose and were overlaid on 0.6% agarose. Medium (0.5 ml) was added to the plates every four days, and colony formation was quantified after fixation and staining with methylene blue after three weeks.
Apoptosis Assay
Apoptosis was measured by annexin staining. Control or infected cells were incubated with annexin-PE antibody (BD Pharmingen, San Diego, Calif.) and counterstained with 7-amino-actinomycin D (7-AAD) per the manufacturer's protocol (BD Pharmingen). Cell fluorescence was measured on a FACScan flow cytometer (BD Pharmingen) and analyzed with Cell Quest software (BD Pharmingen).
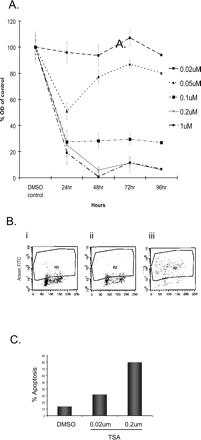
Effects of TSA on D283 cells. (A) Dose-response curves of cell viability for D283 medulloblastoma cells treated with increasing doses of the HDAC inhibitor TSA. (B) FACs analysis of terminal dUTP nick-end-labeled (BD Biosciences) D283 cells treated with TSA for 12 h: FACs dot plots of cells with DMSO (i), and cells treated with 0.02 μM and 0.2 μM TSA (ii and iii). (C) Quantification of FACs dot plots shows that 13% of DMSO-treated cells are apoptotic while 31% in cells exposed to 0.02 μM TSA and 79% in 0.2 μM TSA are apoptotic.
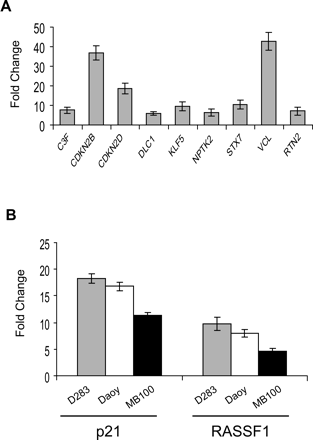
Real-time PCR validation of microarray data. (A) Nine randomly selected genes that were statistically significant as induced at least twofold by TSA on microarray were analyzed by qPCR. Data are shown as fold change of mRNA compared to DMSO-treated (control) D283 cells. All genes were induced by TSA, consistent with the microarray data. (B) Treatment with the HDAC inhibitor TSA (0.2 μM) results in induction of p21 and RASSF1 mRNA in D283 and Daoy medulloblastoma cell lines as well as MB100 primary cells as measured by qPCR (n = 3).
Results
HDAC Inhibition in Medulloblastoma Cells Induces Expression of Genes Involved in Varying Biological Processes
To identify genes regulated by changes in histone H3 K9 acetylation status, we first determined the optimal dose and timing for treating D283 medulloblastoma cells with TSA. The D283 cell line was chosen since it is widely used as a cell model of medulloblastoma and is well characterized (Hallahan et al., 2003; Lawinger et al., 2000). TSA potently decreased D283 medulloblastoma cell viability and induced apoptosis (Fig. 1). For microarray studies, we treated D283 cells with 0.2 μM TSA for 9 h. The dose and time point were chosen based on viability assays and washout experiments (Fig. 1; see also supplementary data, Fig. 1S). At this concentration, cell viability is 100%, but the majority (85%) of cells have committed to cell death by 24 h. In addition, 9 h of TSA exposure results in robust histone acetylation as measured by Western blot analysis (see supplementary data, Fig. 1S). Whole-genome analysis revealed that 714 genes were up-regulated by TSA at least twofold at a maximal statistical stringency (q < 0.001). To confirm the microarray analysis, real-time quantitative PCR (qPCR) was performed on nine randomly selected genes (Fig. 2A). We then demonstrated that the effects of TSA on induction of gene expression are operative in additional medulloblastoma cell lines. TSA treatment induced expression of p21 and RASSF1 in D283 and Daoy medulloblastoma cell lines and in MB100 primary cell cultures. Both p21 and RASSF1 have been previously identified as genes induced by TSA (Hesson et al., 2004; Sowa et al., 1997). We next analyzed the functional significance of the up-regulated genes by mapping them to various pathways using the PANTHER classification system (www.pantherdb.org). Of the 714 genes up-regulated at least twofold, 106 mapped to 68 known signaling pathways (Table 1). Predominant in these were pathways involved in carcinogenesis such as angiogenesis, apoptosis, and more specifically, the Ras, p53, and Wnt signaling cascades. While many of the genes have not been previously associated with medulloblastoma, pathways known to be involved in medulloblastoma pathogenesis, such as sonic hedgehog signaling, as well as EGF and IGF receptor tyrosine kinase signaling, were also identified by the PANTHER analysis. In addition, many TSA-induced genes function in cerebellar development or possibly in medulloblastoma pathogenesis (Table 2). For example, PAX family gene expression has previously been associated with medulloblastoma (Kozmik et al., 1995). Similarly, Notch-mediated signaling was recently associated with tumor formation in medulloblastoma mouse models (Hallahan et al., 2004).
PANTHER pathway analysis mapped 70 genes with altered expression after TSA treatment to known signaling pathways important in cell proliferation and carcinogenesis
| Pathway . | Number of Genes* . | Pathway . | Number of Genes* . |
|---|---|---|---|
| Adrenaline and noradrenaline biosynthesis | 3 | Insulin/IGF pathway-protein kinase B signaling cascade | 3 |
| Alpha adrenergic receptor signaling pathway | 2 | Integrin signaling pathway | 7 |
| Alzheimer disease—amyloid secretase pathway | 1 | Interferon-gamma signaling pathway | 1 |
| Alzheimer disease—presenilin pathway | 2 | Interleukin signaling pathway | 6 |
| Aminobutyrate degradation | 2 | Ionotropic glutamate receptor pathway | 2 |
| Angiogenesis | 14 | JAK/STAT signaling pathway | 1 |
| Apoptosis signaling pathway | 6 | Metabotropic glutamate receptor group II pathway | 1 |
| Axon guidance mediated by netrin | 6 | Metabotropic glutamate receptor group III pathway | 2 |
| Axon guidance mediated by semaphorins | 4 | Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 1 |
| Axon guidance mediated by Slit/Robo | 1 | ||
| B-cell activation | 5 | Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 1 |
| Blood coagulation | 2 | ||
| Cadherin signaling pathway | 4 | Nicotinic acetylcholine receptor signaling pathway | 2 |
| Circadian clock system | 1 | Notch signaling pathway | 2 |
| Cortocotropin-releasing factor receptor-signaling pathway | 3 | O-antigen biosynthesis | 2 |
| Cytoskeletal regulation by rho GTPase | 3 | Ornithine degradation | 1 |
| D1/D5 dopamine receptor-mediated signaling pathway | 4 | Oxidative stress response | 3 |
| D2/D3/D4 dopamine receptor-mediated signaling pathway | 4 | p53 pathway | 7 |
| EGF receptor signaling pathway | 2 | p53 pathway feedback loops 2 | 2 |
| Endothelin signaling pathway | 4 | Parkinson disease | 7 |
| Enkephalin release | 3 | PDGF signaling pathway | 7 |
| FAS signaling pathway | 1 | Pentose phosphate pathway | 2 |
| FGF signaling pathway | 3 | Phenylethylamine degradation | 1 |
| Folate biosynthesis | 1 | PI3 kinase pathway | 2 |
| General transcription by RNA polymerase I | 1 | Ras pathway | 4 |
| Glycolysis | 2 | T-cell activation | 6 |
| Hedgehog signaling pathway | 1 | TGF-beta signaling pathway | 3 |
| Heterotrimeric G-protein signaling pathway-Gi alpha-and Gs alpha-mediated pathway | 11 | Toll receptor signaling pathway | 1 |
| Heterotrimeric G-protein signaling pathway-Gq alpha-and Go alpha-mediated pathway | 9 | Transcription regulation by bZIP transcription factor | 2 |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 4 | VEGF signaling pathway | 3 |
| Wnt signaling pathway | 9 | ||
| 5HT1 type receptor-mediated signaling pathway | 4 | ||
| 5HT2 type receptor-mediated signaling pathway | 5 | ||
| Huntington disease | 4 | 5HT3 type receptor-mediated signaling pathway | 1 |
| Hypoxia response via HIF activation | 3 | 5HT4 type receptor-mediated signaling pathway | 3 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 15 | 5-Hydroxytryptamine degradation | 1 |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 1 | Unclassified | 608 |
| Pathway . | Number of Genes* . | Pathway . | Number of Genes* . |
|---|---|---|---|
| Adrenaline and noradrenaline biosynthesis | 3 | Insulin/IGF pathway-protein kinase B signaling cascade | 3 |
| Alpha adrenergic receptor signaling pathway | 2 | Integrin signaling pathway | 7 |
| Alzheimer disease—amyloid secretase pathway | 1 | Interferon-gamma signaling pathway | 1 |
| Alzheimer disease—presenilin pathway | 2 | Interleukin signaling pathway | 6 |
| Aminobutyrate degradation | 2 | Ionotropic glutamate receptor pathway | 2 |
| Angiogenesis | 14 | JAK/STAT signaling pathway | 1 |
| Apoptosis signaling pathway | 6 | Metabotropic glutamate receptor group II pathway | 1 |
| Axon guidance mediated by netrin | 6 | Metabotropic glutamate receptor group III pathway | 2 |
| Axon guidance mediated by semaphorins | 4 | Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 1 |
| Axon guidance mediated by Slit/Robo | 1 | ||
| B-cell activation | 5 | Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 1 |
| Blood coagulation | 2 | ||
| Cadherin signaling pathway | 4 | Nicotinic acetylcholine receptor signaling pathway | 2 |
| Circadian clock system | 1 | Notch signaling pathway | 2 |
| Cortocotropin-releasing factor receptor-signaling pathway | 3 | O-antigen biosynthesis | 2 |
| Cytoskeletal regulation by rho GTPase | 3 | Ornithine degradation | 1 |
| D1/D5 dopamine receptor-mediated signaling pathway | 4 | Oxidative stress response | 3 |
| D2/D3/D4 dopamine receptor-mediated signaling pathway | 4 | p53 pathway | 7 |
| EGF receptor signaling pathway | 2 | p53 pathway feedback loops 2 | 2 |
| Endothelin signaling pathway | 4 | Parkinson disease | 7 |
| Enkephalin release | 3 | PDGF signaling pathway | 7 |
| FAS signaling pathway | 1 | Pentose phosphate pathway | 2 |
| FGF signaling pathway | 3 | Phenylethylamine degradation | 1 |
| Folate biosynthesis | 1 | PI3 kinase pathway | 2 |
| General transcription by RNA polymerase I | 1 | Ras pathway | 4 |
| Glycolysis | 2 | T-cell activation | 6 |
| Hedgehog signaling pathway | 1 | TGF-beta signaling pathway | 3 |
| Heterotrimeric G-protein signaling pathway-Gi alpha-and Gs alpha-mediated pathway | 11 | Toll receptor signaling pathway | 1 |
| Heterotrimeric G-protein signaling pathway-Gq alpha-and Go alpha-mediated pathway | 9 | Transcription regulation by bZIP transcription factor | 2 |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 4 | VEGF signaling pathway | 3 |
| Wnt signaling pathway | 9 | ||
| 5HT1 type receptor-mediated signaling pathway | 4 | ||
| 5HT2 type receptor-mediated signaling pathway | 5 | ||
| Huntington disease | 4 | 5HT3 type receptor-mediated signaling pathway | 1 |
| Hypoxia response via HIF activation | 3 | 5HT4 type receptor-mediated signaling pathway | 3 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 15 | 5-Hydroxytryptamine degradation | 1 |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 1 | Unclassified | 608 |
The number of affected genes involved in each pathway.
PANTHER pathway analysis mapped 70 genes with altered expression after TSA treatment to known signaling pathways important in cell proliferation and carcinogenesis
| Pathway . | Number of Genes* . | Pathway . | Number of Genes* . |
|---|---|---|---|
| Adrenaline and noradrenaline biosynthesis | 3 | Insulin/IGF pathway-protein kinase B signaling cascade | 3 |
| Alpha adrenergic receptor signaling pathway | 2 | Integrin signaling pathway | 7 |
| Alzheimer disease—amyloid secretase pathway | 1 | Interferon-gamma signaling pathway | 1 |
| Alzheimer disease—presenilin pathway | 2 | Interleukin signaling pathway | 6 |
| Aminobutyrate degradation | 2 | Ionotropic glutamate receptor pathway | 2 |
| Angiogenesis | 14 | JAK/STAT signaling pathway | 1 |
| Apoptosis signaling pathway | 6 | Metabotropic glutamate receptor group II pathway | 1 |
| Axon guidance mediated by netrin | 6 | Metabotropic glutamate receptor group III pathway | 2 |
| Axon guidance mediated by semaphorins | 4 | Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 1 |
| Axon guidance mediated by Slit/Robo | 1 | ||
| B-cell activation | 5 | Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 1 |
| Blood coagulation | 2 | ||
| Cadherin signaling pathway | 4 | Nicotinic acetylcholine receptor signaling pathway | 2 |
| Circadian clock system | 1 | Notch signaling pathway | 2 |
| Cortocotropin-releasing factor receptor-signaling pathway | 3 | O-antigen biosynthesis | 2 |
| Cytoskeletal regulation by rho GTPase | 3 | Ornithine degradation | 1 |
| D1/D5 dopamine receptor-mediated signaling pathway | 4 | Oxidative stress response | 3 |
| D2/D3/D4 dopamine receptor-mediated signaling pathway | 4 | p53 pathway | 7 |
| EGF receptor signaling pathway | 2 | p53 pathway feedback loops 2 | 2 |
| Endothelin signaling pathway | 4 | Parkinson disease | 7 |
| Enkephalin release | 3 | PDGF signaling pathway | 7 |
| FAS signaling pathway | 1 | Pentose phosphate pathway | 2 |
| FGF signaling pathway | 3 | Phenylethylamine degradation | 1 |
| Folate biosynthesis | 1 | PI3 kinase pathway | 2 |
| General transcription by RNA polymerase I | 1 | Ras pathway | 4 |
| Glycolysis | 2 | T-cell activation | 6 |
| Hedgehog signaling pathway | 1 | TGF-beta signaling pathway | 3 |
| Heterotrimeric G-protein signaling pathway-Gi alpha-and Gs alpha-mediated pathway | 11 | Toll receptor signaling pathway | 1 |
| Heterotrimeric G-protein signaling pathway-Gq alpha-and Go alpha-mediated pathway | 9 | Transcription regulation by bZIP transcription factor | 2 |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 4 | VEGF signaling pathway | 3 |
| Wnt signaling pathway | 9 | ||
| 5HT1 type receptor-mediated signaling pathway | 4 | ||
| 5HT2 type receptor-mediated signaling pathway | 5 | ||
| Huntington disease | 4 | 5HT3 type receptor-mediated signaling pathway | 1 |
| Hypoxia response via HIF activation | 3 | 5HT4 type receptor-mediated signaling pathway | 3 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 15 | 5-Hydroxytryptamine degradation | 1 |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 1 | Unclassified | 608 |
| Pathway . | Number of Genes* . | Pathway . | Number of Genes* . |
|---|---|---|---|
| Adrenaline and noradrenaline biosynthesis | 3 | Insulin/IGF pathway-protein kinase B signaling cascade | 3 |
| Alpha adrenergic receptor signaling pathway | 2 | Integrin signaling pathway | 7 |
| Alzheimer disease—amyloid secretase pathway | 1 | Interferon-gamma signaling pathway | 1 |
| Alzheimer disease—presenilin pathway | 2 | Interleukin signaling pathway | 6 |
| Aminobutyrate degradation | 2 | Ionotropic glutamate receptor pathway | 2 |
| Angiogenesis | 14 | JAK/STAT signaling pathway | 1 |
| Apoptosis signaling pathway | 6 | Metabotropic glutamate receptor group II pathway | 1 |
| Axon guidance mediated by netrin | 6 | Metabotropic glutamate receptor group III pathway | 2 |
| Axon guidance mediated by semaphorins | 4 | Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 1 |
| Axon guidance mediated by Slit/Robo | 1 | ||
| B-cell activation | 5 | Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 1 |
| Blood coagulation | 2 | ||
| Cadherin signaling pathway | 4 | Nicotinic acetylcholine receptor signaling pathway | 2 |
| Circadian clock system | 1 | Notch signaling pathway | 2 |
| Cortocotropin-releasing factor receptor-signaling pathway | 3 | O-antigen biosynthesis | 2 |
| Cytoskeletal regulation by rho GTPase | 3 | Ornithine degradation | 1 |
| D1/D5 dopamine receptor-mediated signaling pathway | 4 | Oxidative stress response | 3 |
| D2/D3/D4 dopamine receptor-mediated signaling pathway | 4 | p53 pathway | 7 |
| EGF receptor signaling pathway | 2 | p53 pathway feedback loops 2 | 2 |
| Endothelin signaling pathway | 4 | Parkinson disease | 7 |
| Enkephalin release | 3 | PDGF signaling pathway | 7 |
| FAS signaling pathway | 1 | Pentose phosphate pathway | 2 |
| FGF signaling pathway | 3 | Phenylethylamine degradation | 1 |
| Folate biosynthesis | 1 | PI3 kinase pathway | 2 |
| General transcription by RNA polymerase I | 1 | Ras pathway | 4 |
| Glycolysis | 2 | T-cell activation | 6 |
| Hedgehog signaling pathway | 1 | TGF-beta signaling pathway | 3 |
| Heterotrimeric G-protein signaling pathway-Gi alpha-and Gs alpha-mediated pathway | 11 | Toll receptor signaling pathway | 1 |
| Heterotrimeric G-protein signaling pathway-Gq alpha-and Go alpha-mediated pathway | 9 | Transcription regulation by bZIP transcription factor | 2 |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 4 | VEGF signaling pathway | 3 |
| Wnt signaling pathway | 9 | ||
| 5HT1 type receptor-mediated signaling pathway | 4 | ||
| 5HT2 type receptor-mediated signaling pathway | 5 | ||
| Huntington disease | 4 | 5HT3 type receptor-mediated signaling pathway | 1 |
| Hypoxia response via HIF activation | 3 | 5HT4 type receptor-mediated signaling pathway | 3 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 15 | 5-Hydroxytryptamine degradation | 1 |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 1 | Unclassified | 608 |
The number of affected genes involved in each pathway.
TSA-induced genes in D283 cells with potential roles in medulloblastoma pathogenesis
| Gene . | Biological Function . |
|---|---|
| Notch1 | Neural differentiation |
| WIF1 | Wnt signaling/developmental processes |
| DKK1 | Wnt signaling/developmental processes |
| sFRP1 | Wnt signaling/developmental processes |
| OLIG2 | mRNA transcription regulation |
| MYBL1 | Inhibition of apoptosis/cell cycle control |
| CCNB3 | Cell cycle control/cell proliferation and differentiation |
| LNK | Receptor protein tyrosine kinase/calcium signaling |
| FBOXO33 | Transcription factor |
| NDRG4 | Cell proliferation and differentiation |
| MAK | Protein phosphorylation |
| PAX6 | mRNA transcription regulation/neurogenesis |
| NCAM1 | Cell adhesion-mediated signaling |
| ABCC2 | Small molecule transport/detoxification |
| TMEFF2 | Oncogenesis |
| TPMT | Drug detoxification |
| CTGF | Receptor protein tyrosine kinase signaling pathway |
| INPP5F | Phospholipid metabolism |
| NRXN3 | Cell adhesion-mediated signaling |
| CYR61 | Angiogenesis/cell cycle control |
| Gene . | Biological Function . |
|---|---|
| Notch1 | Neural differentiation |
| WIF1 | Wnt signaling/developmental processes |
| DKK1 | Wnt signaling/developmental processes |
| sFRP1 | Wnt signaling/developmental processes |
| OLIG2 | mRNA transcription regulation |
| MYBL1 | Inhibition of apoptosis/cell cycle control |
| CCNB3 | Cell cycle control/cell proliferation and differentiation |
| LNK | Receptor protein tyrosine kinase/calcium signaling |
| FBOXO33 | Transcription factor |
| NDRG4 | Cell proliferation and differentiation |
| MAK | Protein phosphorylation |
| PAX6 | mRNA transcription regulation/neurogenesis |
| NCAM1 | Cell adhesion-mediated signaling |
| ABCC2 | Small molecule transport/detoxification |
| TMEFF2 | Oncogenesis |
| TPMT | Drug detoxification |
| CTGF | Receptor protein tyrosine kinase signaling pathway |
| INPP5F | Phospholipid metabolism |
| NRXN3 | Cell adhesion-mediated signaling |
| CYR61 | Angiogenesis/cell cycle control |
TSA-induced genes in D283 cells with potential roles in medulloblastoma pathogenesis
| Gene . | Biological Function . |
|---|---|
| Notch1 | Neural differentiation |
| WIF1 | Wnt signaling/developmental processes |
| DKK1 | Wnt signaling/developmental processes |
| sFRP1 | Wnt signaling/developmental processes |
| OLIG2 | mRNA transcription regulation |
| MYBL1 | Inhibition of apoptosis/cell cycle control |
| CCNB3 | Cell cycle control/cell proliferation and differentiation |
| LNK | Receptor protein tyrosine kinase/calcium signaling |
| FBOXO33 | Transcription factor |
| NDRG4 | Cell proliferation and differentiation |
| MAK | Protein phosphorylation |
| PAX6 | mRNA transcription regulation/neurogenesis |
| NCAM1 | Cell adhesion-mediated signaling |
| ABCC2 | Small molecule transport/detoxification |
| TMEFF2 | Oncogenesis |
| TPMT | Drug detoxification |
| CTGF | Receptor protein tyrosine kinase signaling pathway |
| INPP5F | Phospholipid metabolism |
| NRXN3 | Cell adhesion-mediated signaling |
| CYR61 | Angiogenesis/cell cycle control |
| Gene . | Biological Function . |
|---|---|
| Notch1 | Neural differentiation |
| WIF1 | Wnt signaling/developmental processes |
| DKK1 | Wnt signaling/developmental processes |
| sFRP1 | Wnt signaling/developmental processes |
| OLIG2 | mRNA transcription regulation |
| MYBL1 | Inhibition of apoptosis/cell cycle control |
| CCNB3 | Cell cycle control/cell proliferation and differentiation |
| LNK | Receptor protein tyrosine kinase/calcium signaling |
| FBOXO33 | Transcription factor |
| NDRG4 | Cell proliferation and differentiation |
| MAK | Protein phosphorylation |
| PAX6 | mRNA transcription regulation/neurogenesis |
| NCAM1 | Cell adhesion-mediated signaling |
| ABCC2 | Small molecule transport/detoxification |
| TMEFF2 | Oncogenesis |
| TPMT | Drug detoxification |
| CTGF | Receptor protein tyrosine kinase signaling pathway |
| INPP5F | Phospholipid metabolism |
| NRXN3 | Cell adhesion-mediated signaling |
| CYR61 | Angiogenesis/cell cycle control |
DKK1 Is Down-regulated in Medulloblastoma and Induced by HDAC Inhibition
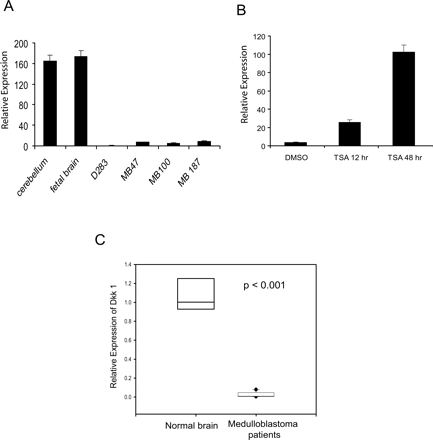
Our goal was to identify genes epigenetically silenced by histone deacetylation that are reversibly induced by TSA and thus are candidate tumor suppressor genes. Of 714 genes up-regulated on TSA treatment, we found several genes previously shown to suppress tumor growth in other cancers. Among these genes was DKK1, a Wnt antagonist that affects cell growth. We examined changes in DKK1 expression on TSA treatment in three patient-derived primary medulloblastoma cell lines (MB47, MB100, and MB187) and one immortalized cell line (D283) with respect to normal cerebellum by reverse transcriptase (RT)-PCR. DKK1 expression was significantly down-regulated in all cases and increased on TSA treatment (Fig. 3A and B).
DKK1 expression is decreased in medulloblastoma cell lines and patients. (A) DKK1 mRNA, determined by qPCR, is significantly decreased in D283 cells and primary medulloblastoma cell cultures compared to normal human cerebellum (n = 3). (B) TSA (0.2 μM) potently induces DKK1 expression in D283 cells as determined by qPCR. (C) Box plot representation of analysis of variance analysis shows significant decrease in DKK1 expression (P < 0.001) in medulloblastoma patients (n = 10) compared to normal cerebellum (n = 3).
To extend these findings to medulloblastoma tumors, we compared DKK1 expression in 10 patient tissue samples relative to normal cerebellum by RT-PCR. When compared to normal cerebellum, all 10 samples expressed 80% less DKK1 (Fig. 3C). Analysis of variance confirmed that this difference was statistically significant (P < 0.001).
Histone Acetylation Regulates DKK1 Expression in Medulloblastoma
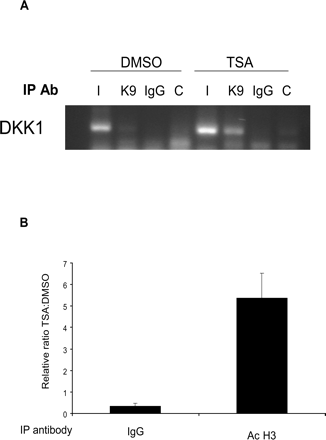
To further validate the role of histone tail modifications as an epigenetic silencing mechanism for DKK1 in medulloblastoma, we performed ChIP using antibodies against acetylated histones H3 at the Lys9 position. Consistent with our earlier results, TSA treatment increased fivefold the histone acetylation in the promoter region of DKK1 (Fig. 4).
These data suggest that reversal of histone deacetylation by TSA was sufficient to allow DKK1 gene expression in medulloblastoma cells.
DKK1 Suppresses Medulloblastoma Growth and Induces Apoptosis
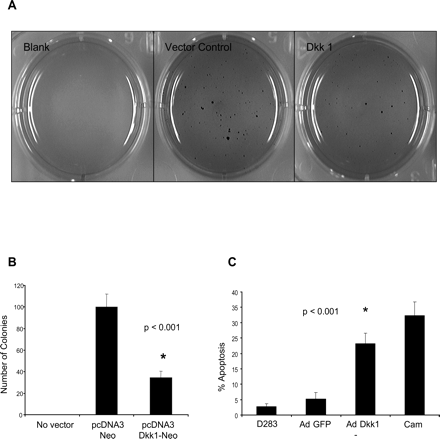
To test whether DKK1 can function as a tumor suppressor in medulloblastoma cells, its effect on growth was measured in colony focus-forming assays. Expression vectors were constructed that expressed the neomycin (neo) resistance gene along with DKK1. Vectors were transfected into D283 cells, selected in neo, and plated onto soft agar. DKK1 expression was confirmed by qPCR measurement of mRNA in control and DKK1-transfected cells (supplemental data, Fig. 2S, panel A). After 3 weeks, cells expressing DKK1 formed 60% fewer neo-resistant colonies than did controls (P < 0.001) (Fig. 5A and B).
Chromatin immunoprecipitation analysis of the DKK1 promoter. (A) Histone-DNA complexes from DMSO- or TSA-treated D283 cells were immunoprecipitated with IgG or antiacetyl-histone (K9) antibodies and amplified with DKK1 primers. Acetyl histone (K9) associated with DKK1 in TSA-treated but not control (DMSO-treated) cells. I, input control; K9, acetyl-histone; IgG, isotype control. (B) Densitometric quantification of ChIP analysis: fivefold increase in DKK1 promoter association with acetylated histone in TSA-treated cells compared to DMSO-treated controls (P < 0.001).
We next tested whether DKK1 expression suppressed tumor development by growth inhibition or induction of tumor cell death. D283 cells were transduced with vectors expressing DKK1, and cell-cycle progression was assayed. Efficiency of Ad-DKK1 infection was evaluated by green fluorescent protein (GFP) fluorescence, and expression was verified by qPCR (supplementary data, Fig. 2S, panels B and C). Ectopically expressing DKK1 did not affect cell cycle kinetics, suggesting that DKK1-inhibited growth did not occur via a block in cell-cycle progression (data not shown). In contrast, DKK1 enhanced apoptosis fourfold in medulloblastoma cells as measured by annexin staining (Fig. 5C). These data support the hypothesis that DKK1 acts as a tumor suppressor gene in medulloblastoma.
Discussion
Epigenetic silencing of tumor suppressor genes controls various aspects of carcinogenesis, including proliferation, differentiation, and apoptosis (Momparler, 2003). This widespread mechanism has been implicated in regulating critical signaling cascades, including Notch, sonic hedgehog, and Wnt (Jones and Baylin, 2002). Aberrant silencing of tumor suppressor genes has been associated with methylation of their promoter regions in medulloblastoma (Lindsey et al., 2005). Little is known, however, about how epigenetic histone modifications may alter gene expression in medulloblastoma. Using D283 cells, a well-characterized medulloblastoma cell line, we examined global epigenetic changes in medulloblastoma and identified genes belonging to multiple pathways important in tumorigenesis. Similar approaches in tumor cell lines by us and others have yielded several promising candidate tumor suppressor genes (Foltz et al., 2006; Suzuki et al., 2002; Takai et al., 2005; Yamashita et al., 2002). In the present screen, we identified DKK1, a Wnt signaling antagonist, and confirmed its silencing in medulloblastoma cell lines, primary tumor cells, and medulloblastoma patient tissue.
The Wnt signaling pathway regulates multiple processes in development, tissue homeostasis, and stem cell maintenance (Nusse, 2005). Genetic mutations that disrupt Wnt signaling can cause tumors, the best-studied case being colon adenocarcinoma (Suzuki et al., 2004). Although mutations in Wnt signaling components, APC, GSK3β, and β-catenin have all been linked to colon cancer progression, mutations in these molecules occur only in a small subset of medulloblastoma patients (Koch et al., 2001), with most being the APC mutations in Turcot's syndrome (Marino, 2005). Our work demonstrates that Wnt signaling is also disrupted in medulloblastoma pathogenesis via the epigenetic silencing of DKK1.
We demonstrated that restoring DKK1 expression in medulloblastoma cells induced apoptosis and suppressed colony formation. Consistent with our data, others showed that expressing DKK1 in HeLa cells also suppressed transformation (Lee et al., 2004; Mikheev et al., 2004), and similar to our results, DKK1 inhibited growth by inducing apoptosis, not cell cycle arrest (Lee et al., 2004). In gliomas as well as models of ischemic neuronal apoptosis, DKK1 was also shown to be a pro-apoptotic factor (Cappuccio et al., 2005; Shou et al., 2002). Thus, DKK1's tumor-suppressing activity is likely important in regulating proliferation in many cell types.
Our data raise two important questions with regard to DKK1 activity in medulloblastoma. The first is how DKK1 induces apoptosis in medulloblastoma. One possibility is that DKK1 suppresses the canonical Wnt signaling pathway, thus down-regulating prosurvival molecules such as Bcl-2. Alternatively, DKK1 might stimulate pro-apoptotic pathways via noncanonical signaling mechanisms. Clues to DKK1 function in medulloblastoma might be provided by its role during vertebrate limb development where DKK1 inhibits proproliferative activities of canonical Wnt signaling and independently regulates apoptosis (Mukhopadhyay et al., 2001). Although the molecular mechanisms that allow DKK1 to regulate apoptosis are not well understood, some data suggest that it regulates the JNK pathway. In mesothelioma, DKK1 antagonizes Wnt signaling in the absence of β-catenin by inducing JNK-mediated apoptosis.
Tumor suppression by DKK1. (A) DKK1-transfected cells produced fewer colonies than control vector on soft agar as visualized by methylene blue staining. (B) Colony counts reveal significantly fewer colonies in DKK1-transfected D283 cell cultures compared to control vector (n = 3, P < 0.001). (C) Ad-mediated DKK1 expression in D283 cells leads to increased apoptosis compared to Ad-GFP controls (n = 3, P < 0.001).
A second question is whether DKK1 is required for medulloblastoma tumor initiation or is associated with tumor progression. Recent evidence from colon cancer supports its role in tumor progression (Aguilera et al., 2006). Investigating DKK1 gene knockdown in mouse models of medulloblastoma will provide insight into its biological role in medulloblastoma tumorigenesis.
In this study, we demonstrated the feasibility and robustness of a systematic approach to determine the role of epigenetically silenced genes in medulloblastoma. Our preliminary data suggest that DKK1 gene is a potent tumor suppressor and that Wnt signaling is important in medulloblastoma pathogenesis, a factor not previously appreciated. We are now investigating the mechanistic basis of DKK1 activity in medulloblastoma. Recent studies indicate that Wnt signaling is negatively regulated by secreted Wnt antagonists such as secreted frizzled-related proteins and Dickkopf proteins. We found Wif1 and sFRP1 also to be silenced in medulloblastoma cell lines and up-regulated on HDAC inhibition by TSA (data not shown). A systematic approach aimed to elucidate molecular mechanisms that various Wnt antagonists use to induce apoptosis in medulloblastoma may indicate new, more effective therapeutic targets. Similarly, studies with other epigenetically silenced genes will delineate their roles in malignant transformation and identify pathways involved in tumorigenesis.
This work was supported by the Departments of Pediatrics and Neurosurgery, University of Iowa College of Medicine, and Carver Foundation grant 99-30.
References
Aguilera, O., Fraga, M.F., Ballestar, E., Paz, M.F., Herranz, M., Espada, J., Garcia, J.M., Munoz, A., Esteller, M., and Gonzalez-Sancho, J.M. (
Bachman, K.E., Park, B.H., Rhee, I., Rajagopalan, H., Herman, J.G., Baylin, S.B., Kinzler, K.W., and Vogelstein, B. (
Cappuccio, I., Calderone, A., Busceti, C.L., Biagioni, F., Pontarelli, F., Bruno, V., Storto, M., Terstappen, G.T., Gaviraghi, G., Fornai, F., Battaglia, G., Melchiorri, D., Zukin, S., Nicoletti, F., and Caricasole, A. (
Foltz, G., Ryu, G.Y., Yoon, J.G., Nelson, T., Fahey, J., Frakes, A., Lee, H., Field, L., Zander, K., Sibenaller, Z., Ryken, T.C., Vibhakar, R., Hood, L., and Madan, A. (
Fruhwald, M.C., O'Dorisio, M.S., Dai, Z., Tanner, S.M., Balster, D.A., Gao, X., Wright, F.A., and Plass, C. (
Hallahan, A.R., Pritchard, J.I., Chandraratna, R.A., Ellenbogen, R.G., Geyer, J.R., Overland, R.P., Strand, A.D., Tapscott, S.J., and Olson, J.M. (
Hallahan, A.R., Pritchard, J.I., Hansen, S., Benson, M., Stoeck, J., Hatton, B.A., Russell, T.L., Ellenbogen, R.G., Bernstein, I.D., Beachy, P.A., and Olson, J.M. (
Hesson, L., Bieche, I., Krex, D., Criniere, E., Hoang-Xuan, K., Maher, E.R., and Latif, F. (
Jones, P.A., and Baylin, S.B. (
Koch, A., Waha, A., Tonn, J.C., Sörensen, N., Berthold, F., Wolter, M., Reifenberger, J., Hartmann, W., Friedl, W., Reifenberger, G., Wiestler, O.D., and Pietsch, T. (
Kozmik, Z., Sure, U., Ruedi, D., Busslinger, M., and Aguzzi, A. (
Lawinger, P., Venugopal, R., Guo, Z.S., Immaneni, A., Sengupta, D., Lu, W., Rastelli, L., Marin Dias Carneiro, A., Levin, V., Fuller, G.N., Echelard, Y., and Majumder, S. (
Lee, A.Y., He, B., You, L., Xu, Z., Mazieres, J., Reguart, N., Mikami, I., Batra, S., and Jablons, D.M. (
Lindsey, J.C., Lusher, M.E., Anderton, J.A., Bailey, S., Gilbertson, R.J., Pearson, A.D., Ellison, D.W., and Clifford, S.C. (
Lindsey, J.C., Anderton, J.A., Lusher, M.E., and Clifford, S.C. (
MacDonald, T.J., Rood, B.R., Santi, M.R., Vezina, G., Bingaman, K., Cogen, P.H., and Packer, R.J. (
Marino, S. (
Mikheev, A.M., Mikheeva, S.A., Liu, B., Cohen, P., and Zarbl, H. (
Mukhopadhyay, M., Shtrom, S., Rodriguez-Esteban, C., Chen, L., Tsukui, T., Gomer, L., Dorward, D.W., Glinka, A., Grinberg, A., Huang, S.P., Niehrs, C., Belmonte, J.C.I. and Westphal, H. (
Mulhern, R.K., Palmer, S.L., Merchant, T.E., Wallace, D., Kocak, M., Brouwers, P., Krull, K., Chintagumpala, M., Stargatt, R., Ashley, D.M., Tyc, V.L., Kun, L., Boyett, J., and Gajjar, A. (
Packer, R.J., Cogen, P., Vezina, G., and Rorke, L.B. (
Packer, R.J., Rood, B.R., and MacDonald, T.J. (
Rood, B.R., Macdonald, T.J., and Packer, R.J. (
Shou, J., Ali-Osman, F., Multani, A.S., Pathak, S., Fedi, P., and Srivenugopal, K.S. (
Sowa, Y., Orita, T., Minamikawa, S., Nakano, K., Mizuno, T., Nomura, H., and Sakai, T. (
Storey, J.D., and Tibshirani, R. (
Suzuki, H., Gabrielson, E., Chen, W., Anbazhagan, R., van Engeland, M., Weijenberg, M.P., Herman, J.G., and Baylin, S.B. (
Suzuki, H., Watkins, D.N., Jair, K.W., Schuebel, K.E., Markowitz, S.D., Chen, W.D., Pretlow, T.P., Yang, B., Akiyama, Y., van Engeland, M., Toyota, M., Tokino, T., Hinoda, Y., Imai, K., Herman, J.G., and Baylin, S.B. (
Takai, N., Kawamata, N., Walsh, C.S., Gery, S., Desmond, J.C., Whittaker, S., Said, J.W., Popoviciu, L.M., Jones, P.A., Miyakawa, I., and Koeffler, H.P. (