Nutrient acquisition and metabolism by Campylobacter jejuni
- Ottawa Institute of Systems Biology, Department of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Ottawa, ON, Canada
The gastrointestinal pathogen Campylobacter jejuni is able to colonize numerous different hosts and compete against the gut microbiota. To do this, it must be able to efficiently acquire sufficient nutrients from its environment to support its survival and rapid growth in the intestine. However, despite almost 50 years of research, many aspects as to how C. jejuni accomplishes this feat remain poorly understood. C. jejuni lacks many of the common metabolic pathways necessary for the use of glucose, galactose, or other carbohydrates upon which most other microbes thrive. It does however make efficient use of citric acid cycle intermediates and various amino acids. C. jejuni readily uses the amino acids aspartate, glutamate, serine, and proline, with certain strains also possessing additional pathways allowing for the use of glutamine and asparagine. More recent work has revealed that some C. jejuni strains can metabolize the sugar L-fucose. This finding has upset years of dogma that C. jejuni is an asaccharolytic organism. C. jejuni also possesses diverse mechanisms for the acquisition of various transition metals that are required for metabolic activities. In particular, iron acquisition is critical for the formation of iron–sulfur complexes. C. jejuni is also unique in possessing both molybdate and tungsten cofactored proteins and thus has an unusual regulatory scheme for these metals. Together these various metabolic and acquisition pathways help C. jejuni to compete and thrive in wide variety of hosts and environments.
Introduction
Campylobacter jejuni is the most common cause of gastroenteritis in the developed world. The most frequent sources of infection are through contaminated food, particularly poultry products, or through contaminated raw milk or drinking water (Skirrow, 1991). When consumed by its potential host, C. jejuni will pass through the stomach and establish itself in the host intestinal tract. C. jejuni causes gastroenteritis in humans, with infections typically characterized by diarrhea (sometimes bloody), abdominal pain, and fever (Galanis, 2007). However, C. jejuni will colonize to high levels in poultry or other birds without eliciting an immune response. This commensal colonization allows C. jejuni to become established in poultry flocks, which often serve as a reservoir for human infection (Lee and Newell, 2006). Poultry meat is easily contaminated during processing, and if the resulting meat is not properly prepared, C. jejuni can cause infection in humans with a dose as small as a few hundred bacterial cells (Robinson, 1981).
While we have acquired significant knowledge about Campylobacter, especially since its genome was first sequenced in 2000 (Parkhill et al., 2000), many aspects of its biology have remained a mystery. Recent work on C. jejuni has allowed us to better understand how C. jejuni acquires and metabolizes key nutrients (Velayudhan et al., 2004; del Rocio Leon-Kempis et al., 2006; Hofreuter et al., 2008; Stahl et al., 2011), but we are still only beginning to gain a full appreciation for the role these nutrients play in host colonization. To be successful, C. jejuni must possess the ability to survive in a wide variety of hosts, from an avian host, to its pathogenic colonization of a human host. C. jejuni must also be able to survive in the environment, in water or food, before being ingested by potential hosts. This requires a certain degree of metabolic flexibility to make sufficient use of the different metabolites that are available in each niche. The extent of C. jejuni’s metabolic flexibility is particularly impressive given the relatively small number of different metabolic pathways initially identified in its genome (Parkhill et al., 2000).
Carbon and Nitrogen Acquisition and Metabolism
Glycolytic Pathways
Unlike most other bacteria, C. jejuni lacks the ability to use many common carbohydrates as carbon sources (Parkhill et al., 2000). Aside from lacking the proper transporters to take up sugars like glucose or galactose, it also lacks several key enzymes within the glycolytic pathway. Although C. jejuni possesses the complete set of enzymes necessary for the synthesis of glucose-6-phosphate from pyruvate via gluconeogenesis (Velayudhan and Kelly, 2002), it lacks any form of glucokinase to phosphorylate extracellular glucose. C. jejuni is also missing the 6-phosphofructokinase enzyme that is required for the irreversible phosphorylation of fructose-6-phosphate to fructose-1,6-diphosphate during glycolysis. Interestingly, it still possesses pyruvate kinase, an enzyme that is required for another irreversible step of glycolysis, but not gluconeogenesis (Velayudhan and Kelly, 2002). It has been hypothesized that C. jejuni may catabolize some molecules such as glycerol-3-phosphate, which would require the presence of only the final portions of the glycolytic pathway. The use of glycerol-3-phosphate is supported by the presence of a glycerol-3-phosphate transporter (GlpT) in the strain 81–176 (Hofreuter et al., 2006). In addition to this, although C. jejuni possesses the enzymes transaldolase, transketolase, ribulose-3-phosphate epimerase, and ribose-5-phosphate isomerase, which make up the non-oxidative portion of the pentose phosphate pathway, it completely lacks the oxidative portion of the pathway (Velayudhan and Kelly, 2002; Line et al., 2010). This precludes the possibility that it may be able to metabolize pentose sugars through this pathway (Line et al., 2010), but the function of the remaining genes in C. jejuni remains unknown.
The annotation of the C. jejuni genome also failed to identify any additional carbohydrate metabolic pathways, however, both Muraoka and Zhang (2010) and Stahl et al. (2011), independently identified a novel L-fucose pathway present within certain strains of C. jejuni. These reports identified a L-fucose permease (Cj0486/FucP), present within a genomic island encompassing the genes cj0480c–cj0490. This permease has 37.2% amino acid identity to the L-fucose permease identified in E. coli and contains three highly conserved amino acids (Asp46, Glu135, and Asn162), which have been found previously to be critical for L-fucose active transport (Dang et al., 2010). Also contained within this genomic island is a second transporter, Cj0484, annotated as a MFS superfamily efflux pump. However, it does not bear any significant similarities to L-fucose permeases and the function of this transporter remains unknown. Despite strong similarities between the E. coli and C. jejuni permeases themselves, the rest of the pathway in C. jejuni does not resemble the known L-fucose metabolic pathways characterized in E. coli (Baldoma and Aguilar, 1988) or Bacteroides spp. (Coyne et al., 2005). The C. jejuni pathway does however show some similarities to a L-fucose dehydrogenase dependent pathway found in Xanthomonas campestris (Yew et al., 2006; Stahl et al., 2011). Despite these similarities, the precise mechanisms of this pathway remain unknown, and will require further study.
Although this pathway was completely necessary for the uptake and metabolism of L-fucose in vitro, when colonizing the intestine, its utility appears to be host dependent. Chick colonization assays using mutants in the FucP permease did not show any impairment for colonization relative to the wild-type strains. When the same colonization assay was performed in a colostrum-deprived piglet model of infection, a roughly 15- to 100-fold reduction in the mutant relative to wild-type was observed. This reduction relative to the wild-type demonstrates that this pathway is advantageous for colonization of certain hosts, but not completely necessary for colonization (Stahl et al., 2011).
Amino Acids
Among the primary nutrient sources for C. jejuni, are a handful of amino acids that it can acquire while growing within the gut. C. jejuni also will draw on available amino acids in a sequential manner, with serine, aspartate, asparagine, and glutamate being preferred in that order, although this will vary between strains. Proline will also be metabolized, but only after other nutrients have begun to become exhausted (Wright et al., 2009). It should also be noted that serine, aspartate, glutamate, and proline make up some of the most common amino acids found in chick excreta, perhaps explaining why they play such a central role in C. jejuni metabolism (Parsons, 1984).
Serine
Serine metabolism is achieved due to the action of the SdaC (Cj1624c) and SdaA (Cj1625c) proteins (Mendz et al., 1997; Velayudhan et al., 2004). SdaC acts as a transporter for serine into the cell, while SdaA is a L-serine dehydratase. Work by Velayudhan et al. (2004) found that the SdaC transport protein acts as a high capacity, low affinity L-serine transporter. Deletion mutants into this gene completely impaired serine uptake into the cell, indicating that SdaC is the sole transporter capable of transporting significant amounts of L-serine into the cell. Additionally, due to SdaC’s low specific affinity for L-serine, SdaC also exhibits some ability to transport other amino acids into the cell as well, particularly L-threonine, raising the possibility this transporter plays a broader role in amino acid uptake (Velayudhan et al., 2004).
In the same study, the SdaA protein was confirmed to be an L-serine dehydratase and was necessary for the metabolism of L-serine (Velayudhan et al., 2004). Interestingly, unlike most other L-serine dehydratases, the protein found in C. jejuni was not pyridoxal-5′-phosphate dependent, but rather utilized a [4Fe–4S] cluster in a manner more reminiscent of many anaerobic bacteria (Velayudhan et al., 2004). This enzyme is one among many enzymes in key metabolic pathways that contain oxygen sensitive iron–sulfur complexes (Figure 1). It has been proposed that the presence of these iron–sulfur containing enzymes is one of the key reasons for C. jejuni’s sensitivity to oxygen, as molecular oxygen or reactive oxygen species, in sufficient quantity will oxidize and damage the iron–sulfur complexes (Flint et al., 1993).
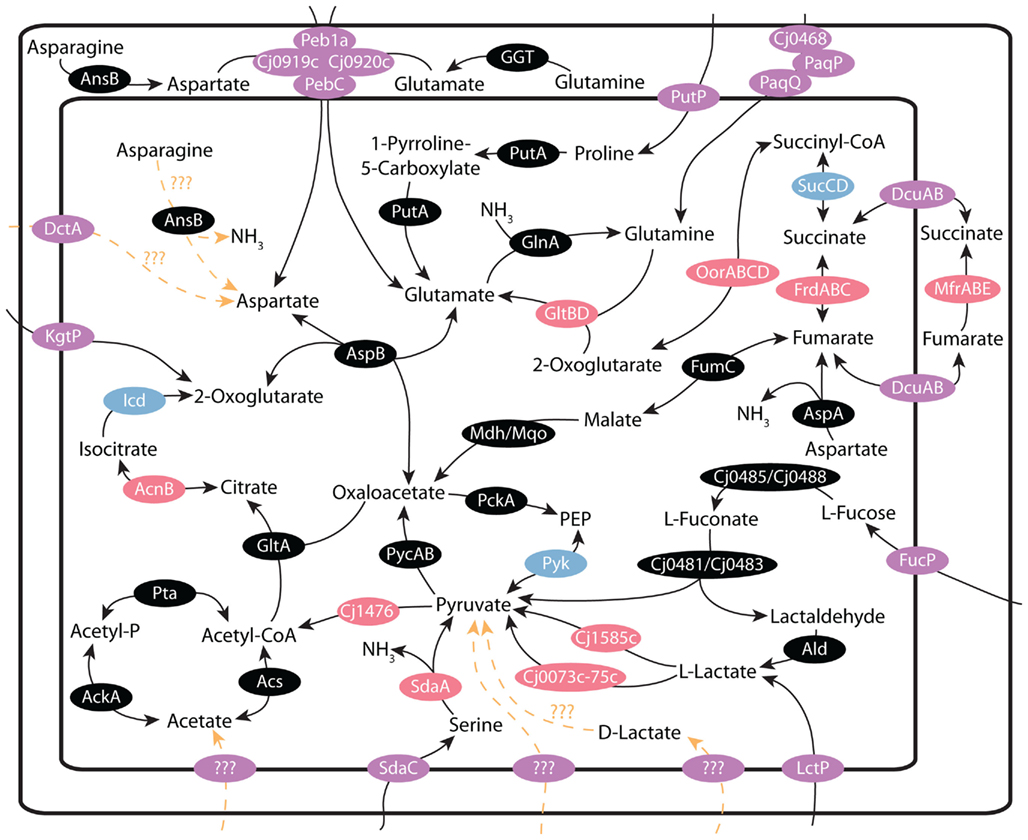
Figure 1. Central metabolism in C. jejuni: schematic of the transporters and anaplerotic reactions that make up the central metabolic pathways of C. jejuni. The function and experimental evidence for each reaction is explained in the text. Note the central role that pyruvate plays in C. jejuni central metabolism; as serine, lactate, and L-fucose metabolism results in the production of pyruvate. Glutamate and aspartate also play key roles in the production of citric acid cycle intermediates. Enzymes or enzyme complexes containing iron–sulfur complexes are indicated in pink, magnesium containing enzymes are indicated in blue, transporters are denoted as purple and hypothetical reactions are indicated in orange.
L-Serine utilization has also been found to be absolutely critical for host colonization, with mutants in SdaC or SdaA being completely impaired for colonization in chicks (Velayudhan et al., 2004). However, these same mutants were able to successfully grow in vitro. This result suggests that while L-serine is not essential for C. jejuni growth, it is necessary in the environment of the gut, where L-serine usage must provide C. jejuni with a key carbon source while establishing itself within the intestinal microbiota (Velayudhan et al., 2004).
Aspartate/glutamate
Aspartate and glutamate metabolism are very similar to each other. Both amino acids are taken up through the Peb1 system, composed of the proteins Cj0919c, Cj0920c, Cj0921c (Peb1A), and Cj0922c (PebC). Work by del Rocio Leon-Kempis et al. (2006) has confirmed the function of Peb1A as the transporter responsible for most, if not all, of the uptake of aspartate and glutamate into the cell. The Peb1a protein itself had been previously identified by Fauchere et al. (1989) as linked to cell adhesion and further work using mutational analysis also found that mutants in Peb1A were significantly impaired for adhesion to epithelial cells (Pei et al., 1998). del Rocio Leon-Kempis et al. (2006) confirmed that this protein was found in the periplasm, and mutants in the gene for Peb1a were mostly impaired for L-aspartate uptake and were completely impaired for L-glutamate uptake. The remaining uptake of L-aspartate in the mutant strain was attributed to the function of the C4-dicarboxylate transporters (DctA, DcuA, and DcuB), but was too minimal to allow for growth utilizing L-aspartate or L-glutamate as a sole carbon source (del Rocio Leon-Kempis et al., 2006).
Once glutamate has been taken up into the cell, the enzyme AspB (Cj0762c), allows for the transamination of glutamate to aspartate. One molecule of glutamate and oxaloacetate are used to produce aspartate and 2-oxoglutarate in a reversible reaction (Guccione et al., 2008). The aspartate can then be deaminated by the aspartase AspA (Cj0087) into fumarate, which can be fed into the citric acid cycle (Guccione et al., 2008).
Proline
Proline has been found to be a less preferred amino acid compared to serine, glutamate, and aspartate, but is still utilized by most strains of C. jejuni (Wright et al., 2009). As a major constituent of mucins, along with L-serine and L-threonine, it is a plentiful nutrient source within the intestine. Proline uptake and metabolism has been linked to the proteins PutP and PutA. PutP (Cj1502c) acts as a sodium/proline symporter, while PutA (Cj1503c) is a bifunctional proline dehydrogenase/delta-1-pyrroline-5-carboxylate dehydrogenase. PutA works to consume L-proline and FAD, producing 1-pyrroline-5-carboxylate, while reducing the electron acceptor FAD to FADH2. PutA subsequently catalyzes the second step of the pathway, by acting as a 1-pyrroline-5-carboxylate dehydrogenase, and oxidizes 1-pyrroline-5-carboxylate to glutamate with the concomitant reduction of NAD to NADH. Glutamate can then be further metabolized to aspartate and subsequently fumarate as described above. Although neither PutP nor PutA has been specifically studied in C. jejuni, they are closely homologous to the well-studied PutA and PutP proteins in E. coli (Tanner, 2008; Zhou et al., 2008) and Helicobacter spp. (Krishnan et al., 2008; Nakajima et al., 2008). In Helicobacter, PutA has been found to be a key factor for stomach colonization and swarming motility (Nakajima et al., 2008). Whether PutA may have similar importance in C. jejuni has yet to be described.
Asparagine
While most strains of C. jejuni possess an asparaginase gene (ansB), C. jejuni does not possess any identifiable asparagine transporters, and indeed many strains that possess ansB (cj0029) are unable to grow on asparagine as a primary carbon source. Nevertheless, certain strains, including 81–176, possess an ansB gene that contains an additional ∼40 bp sequence that encodes for a Sec-dependent secretion signal peptide (Hofreuter et al., 2008). The AnsB protein is normally found exclusively in the cytoplasm, however with this secretion signal peptide, AnsB is transported to the periplasm, where it would be capable of deaminating periplasmic asparagine to aspartate. Thus strains that lack an asparagine transporter can utilize asparagine since the aspartate formed by AnsB can be transported to the cytoplasm by the Peb1 complex and fed to the citric acid cycle (CAC) by AspA (Hofreuter et al., 2008).
Interestingly, it was found that there are two alternate transcriptional start sites in the ansB gene of 81–176, allowing it to produce both the cytoplasmic and periplasmic versions of AnsB. It has been hypothesized by Hofreuter et al. (2008) that the cytoplasmic AnsB might metabolize asparagine originating from small peptides. Although little research has been done to identify small-peptide transporters in C. jejuni, cstA (Cj0917c) has been annotated as a small-peptide transporter and could possibly serve this purpose. Mutants in the asnB gene and/or its signal peptide are still capable of colonizing the intestine in a comparable fashion to the wild-type strain, however, it was found that strains lacking the periplasmic AnsB suffered a significant defect for liver colonization in infected mice. This suggests a tissue specific value for asparagine metabolism (Hofreuter et al., 2008).
Glutamine
The ability of C. jejuni to utilize glutamine has remained somewhat controversial. Based on gene annotations and experimental evidence, C. jejuni contains a functional glutamine uptake system, encoded by the genes cj0467 (paqP), cj0468, and cj0469 (paqQ; Lin et al., 2009). Additionally, C. jejuni contains genes encoding for a glutamate dehydrogenase (gltBD), capable of converting glutamine and 2-oxoglutarate into two molecules of glutamate. Finally, C. jejuni also contains GlnA for the reverse reaction of converting glutamate to glutamine in an ATP dependant manner. This reaction also serves as a means to incorporate free ammonia, a major route of nitrogen assimilation for C. jejuni. However, despite having the biochemical means of transporting and converting glutamine to glutamate via glutamate dehydrogenase, the C. jejuni NCTC11168 strain has never been shown to be able to utilize glutamine as a primary carbon source (Hofreuter et al., 2008), unlike the other previously discussed amino acids. However other C. jejuni strains, such as 81–176 and 81116, contain γ-glutamyl transpeptidase (GGT) and are readily able to use glutamine as a primary carbon source (Hofreuter et al., 2006). GGT is secreted to the periplasm, and acts to hydrolyze either glutamine to glutamate and ammonia, or glutathione to glutamine and a γ-glutamylcysteine. This whole process takes place within the periplasm of the cell. While C. jejuni has not been shown to grow on γ-glutamylcysteine, the Peb1 complex readily takes up the glutamate produced from glutathione or glutamine, where it can be metabolized by AspA and AspB as previously discussed (Hofreuter et al., 2008).
The presence of the GGT enzyme in certain strains appears to have a significant effect on colonization. Knockouts of the GGT enzyme (Cjj_0067) in the 81–176 strain impaired its colonization of MyD88−/− deficient mice (Hofreuter et al., 2008), while a GGT knockout (C8j_0033) in the 81116 strain was defective for chick colonization (Barnes et al., 2007). Although both of these knockouts showed considerable colonization disadvantages relative to their wild-type strains, other strains of C. jejuni lacking GGT are perfectly capable of colonizing these two hosts (Barnes et al., 2007; Hofreuter et al., 2008). This suggests that individual strains may have varying metabolic requirements, making certain pathways dispensable for them. The lack of GGT in a strain such as NCTC11168 may be compensated for the presence of other pathways, such as the recently discovered L-fucose acquisition and metabolic pathway (Stahl et al., 2011). Overall, the diversity of available mechanisms for amino acid acquisition and utilization between strains may allow different C. jejuni strains to co-colonize and to colonize multiple niches or different hosts, thereby expanding the versatility of the species.
Short Chain Fatty Acids
Within the ecosystem of the gut, large amounts of short chain fatty acids (SCFA) are produced as metabolic byproducts from certain bacteria. The host or other bacteria subsequently metabolize these SCFA. One example of this is the SCFA butyrate, which is produced as a byproduct of acetate metabolism in certain bacteria (Duncan et al., 2004). Butyrate is readily taken up by colonocytes and serves as a primary energy source for these cells (Louis et al., 2010). The most significant SCFA found within the gut is acetate (Duncan et al., 2004), with smaller amounts of lactate, propionate, and butyrate also present (Belenguer et al., 2011). Although there are bacteria that will make use of all of these molecules, known pathways and experimental evidence in C. jejuni suggest that it only has the capacity to transport and metabolize acetate and lactate (Wright et al., 2009; Thomas et al., 2011).
Acetate
Acetate is initially produced and secreted as a byproduct in several of C. jejuni’s primary metabolic pathways such as L-serine catabolism. However, in later stages of growth, when more preferable sources are exhausted, acetate can also be used as a carbon source for C. jejuni. Although a transporter for acetate has not been identified in C. jejuni, with the exception of an annotated acetate permease (actP) in C. jejuni subsp. doylei (Parker et al., 2007), it has been experimentally demonstrated in vitro that C. jejuni has the ability to use acetate as a primary carbon source (Wright et al., 2009).
The production and excretion of acetate comes as the result of the need to recycle CoA. The enzyme Pta (Cj0688) produces acetyl-phosphate from acetyl-CoA, freeing the CoA for reuse. The enzyme acetate kinase [AckA (Cj0689)] then produces acetate from acetyl-phosphate, which is excreted and builds up in the surrounding medium. As other sources of nutrients become scarcer, a switch in the direction of the pathway allows for the uptake of acetate into the cell, and the synthesis of acetyl-CoA from acetate by Acetyl-CoA synthase [Acs (Cj1537c)], or by reversing the direction of the AckA–Pta pathway (Velayudhan and Kelly, 2002; Wright et al., 2009).
Lactate
Lactate is also produced in large quantities in the colon as a byproduct of carbohydrate fermentation by numerous varieties of bacteria commonly found in the gut. However, despite its rapid production, overall levels of lactate within the gut remain relatively low, as high rates of production are matched by rapid consumption of lactate by other bacteria (Belenguer et al., 2011). A recent study by Thomas et al. (2011) has well documented the usage of lactate by C. jejuni, although several points still require further research.
While lactate is a common byproduct of fermentation in some bacteria, there is no evidence of lactate excretion by C. jejuni. Moreover, while C. jejuni possesses an annotated L-lactate permease (lctP) a mutant into the lctP gene (cj0076c), was still capable of some L-lactate transport, suggesting the existence of a second, as of yet unidentified lactate permease (Thomas et al., 2011).
Once inside the cell, L-lactate can be catabolized into pyruvate through one of several pathways. The first involves three genes, cj0073c, cj0074c, and cj0075c (lldEFG/lutABC). These three genes code for a non-flavin iron–sulfur containing oxidoreductase complex that demonstrate NAD-independent, L-lactate dehydrogenase activity (L-iLDH), thereby converting L-lactate to pyruvate (Thomas et al., 2011). A second, flavin and iron–sulfur containing enzyme Cj1585c (Dld), also demonstrated L-iLDH activity, although the reason for this redundancy is not immediately clear, and indeed, several strains of C. jejuni such as the 81–176 and 81116 strains, lack homologs to cj1585c. A lactate dehydrogenase (Ldh) was also annotated in the C. jejuni genome; however, Thomas et al. were unable to find a role for this enzyme in lactate metabolism. Knocking out ldh (cj1167) did not impede lactate utilization, while a double knockout in the other two pathways completely eliminated lactate utilization, suggesting that this gene has been either misannotated, or is used under only specific conditions not found in their study (Thomas et al., 2011). C. jejuni has also been found to grow on D-lactate, and demonstrated D-iLDH activity. However, D-lactate metabolism was not significantly affected in the cj0075c or cj1585c mutants and no other transporters or pathways have been identified as responsible for this phenotype (Thomas et al., 2011).
Citric Acid Cycle Intermediates
Campylobacter jejuni depends heavily on the citric acid cycle for its energy needs. All of the aforementioned pathways produce pyruvate, fumarate, oxaloacetate, or 2-oxoglutarate, each of which is fed directly into the citric acid cycle. In addition to this, C. jejuni is capable of transporting several of the citric acid cycle intermediates, and using them directly as nutrient sources. For example 2-oxoglutarate can be transported by the permease KgtP (Cj1619). In addition, succinate, fumarate, and malate can all be transported by the C4-dicarboxylate transporters, DcuA (Cj0088), and DcuB (Cj0671) (Guccione et al., 2008). The putative C4-dicarboxylate transporter DctA may also play a minor role in the uptake of both aspartate and CAC intermediates (del Rocio Leon-Kempis et al., 2006; Guccione et al., 2008); however, this has not been conclusively demonstrated. Additionally, there exists the gene cj0025c, annotated as a putative sodium:dicarboxylate family transmembrane symporter (Parkhill et al., 2000). However, no research has been done to characterize this transporter as of yet, so its possible role as a dicarboxylate transporter has yet to be described. Many strains also possess a citrate transporter (Cj0203 in C. jejuni NCTC11168), allowing for the acquisition of citrate (Parkhill et al., 2000). C. jejuni is known to utilize pyruvate as a primary carbon source while it is present in the growth medium (Velayudhan and Kelly, 2002), however, no pyruvate transporters have been identified or studied in C. jejuni, leaving the mechanism by which it transports pyruvate into the cell currently unknown.
The dicarboxylate transporters DcuA and DcuB may play an important role in C. jejuni thanks to the prominent role of both fumarate and succinate as electron donors and acceptors respectively. C. jejuni possesses the cytoplasmic fumarate reductase FrdABC (Cj0408–Cj0410) that is both capable of oxidizing succinate to fumarate and also catalyzing the reverse reaction. Although C. jejuni was initially annotated as possessing succinate dehydrogenase (sdhABC/Cj0437–Cj439), it was later found that these genes do not encode for succinate dehydrogenase (Weingarten et al., 2009), but rather for a periplasmic methylmenaquinol:fumarate reductase (mfrABE) (Guccione et al., 2010). This complex was found to be in the periplasm, and reduced fumarate, crotonate, and mesaconate to succinate, butyrate, and 2-methylsuccinate respectively under low-oxygen conditions. The cytoplasmic–periplasmic membrane transport of succinate and fumarate necessary for these processes is believed to be facilitated by the DcuA and DcuB transporters (Guccione et al., 2010).
Transition Metal Uptake and Utilization by C. jejuni
In addition to metabolites needed for carbon and energy, the need to have a steady source of micronutrients such as transition metals plays a critical role in the growth of any bacterium, including C. jejuni. Metals play vital and varied roles in proteins and metabolic processes. While some provide purely structural roles in stabilizing protein conformations, many others are the key elements of enzyme catalytic centers. These catalytic centers include the iron–sulfur complexes present within the active sites of numerous enzymes previously mentioned above and in Figure 1. In addition to these previous examples, iron and other metals are critical for the proper functioning of the electron transport chain. Various metals, such as iron and copper, are used as electron donors/acceptors to facilitate electron flow between proteins.
A complete understanding of the roles played by transition metals is still in its infancy in C. jejuni. However the information gleaned thus far indicates that transition metal uptake and utilization are important factors for C. jejuni growth, colonization, and virulence.
Iron
The importance of iron acquisition to the cell cannot be understated. In the absence of sufficient iron sources, the growth of C. jejuni is significantly reduced, if not completely eliminated (Palyada et al., 2004). It is this importance that has led C. jejuni and other bacteria to develop a wide variety of mechanisms for iron acquisition; the details of which have been extensively discussed elsewhere and will only be summarized in this review (Stintzi et al., 2008; Miller et al., 2009; Butcher et al., 2010).
One of the key roles for iron within C. jejuni is as part of iron–sulfur complexes. These complexes exist in the active sites of several key enzymes within the metabolic networks outlined in Figure 1. These include SdaA, AcnB, Cj1585c, Cj0074c, FrdB, MfrB, GltD, OorD, and Cj1476c, plus many more not discussed in this review. It is important to note, that these proteins occupy key points within C. jejuni metabolism, and involve the electron transport chain, serine metabolism, L-lactate metabolism, as well as the citric acid cycle. Without these proteins, many of C. jejuni’s key metabolic pathways would cease to function, explaining why iron acquisition is critical to C. jejuni colonization and growth. The presence of enzymes containing iron–sulfur complexes at key points in C. jejuni’s metabolic pathways also contributes to C. jejuni sensitivity to high levels of oxygen, since the iron–sulfur complexes are sensitive to damage by both molecular oxygen, and reactive oxygen species (Flint et al., 1993).
The C. jejuni FeoB (Cj1398) specifically imports ferrous ions across the inner membrane of the cell; and ferrous ion uptake is an important factor for colonization of the chick gastrointestinal tract. In fact, a ΔfeoB mutant in the NCTC11168 strain showed more than a 104 fold reduction in chick colonization (Naikare et al., 2006). C. jejuni also contains a FeoA homolog (Cj1397), but its role in ferrous ion import is currently undefined (van Vliet et al., 2002). A small fraction of ferrous ion import could also traffic through ZupT (Cj0263), a transporter primarily responsible for zinc uptake (Figure 2). The E. coli ZupT homolog has been shown to possess a broad substrate affinity for divalent cations including ferrous ions in E. coli, however, it has yet to be determined if this remains true in the case of C. jejuni (Grass et al., 2005). It should however be noted that iron will generally not be found in a free form inside a living organism, even in the low-oxygen environment of the intestinal tract. Iron will generally be bound in complex with various molecules such as organic acids (citric, succinic, malic acids) and amino acids (asparagine, glycine; Butcher et al., 2010). Iron can also be present in prosthetic groups such as heme. Hosts also restrict the amount of iron available to the microbiota by secreting iron-binding proteins such as transferrin or lactoferrin. In response to this host restriction, the microbiota secretes siderophores (e.g., enterobactin) to acquire the iron needed for proper growth. Thus pathogens such as C. jejuni typically possess pathways for importing several of these diverse iron sources. C. jejuni has pathways for the import of iron containing complexes such as ferric–enterobactin (CeuBCDE/CfrA/CfrB; Palyada et al., 2004; Xu et al., 2010), heme (ChuABCDZ; Ridley et al., 2006), ferric–rhodotorulic acid (p19/Cj1658-63; Miller et al., 2009; Butcher et al., 2010), and ferric–transferrin/lactoferrin (CfbpABC/CtuA; Miller et al., 2008; Figure 2).
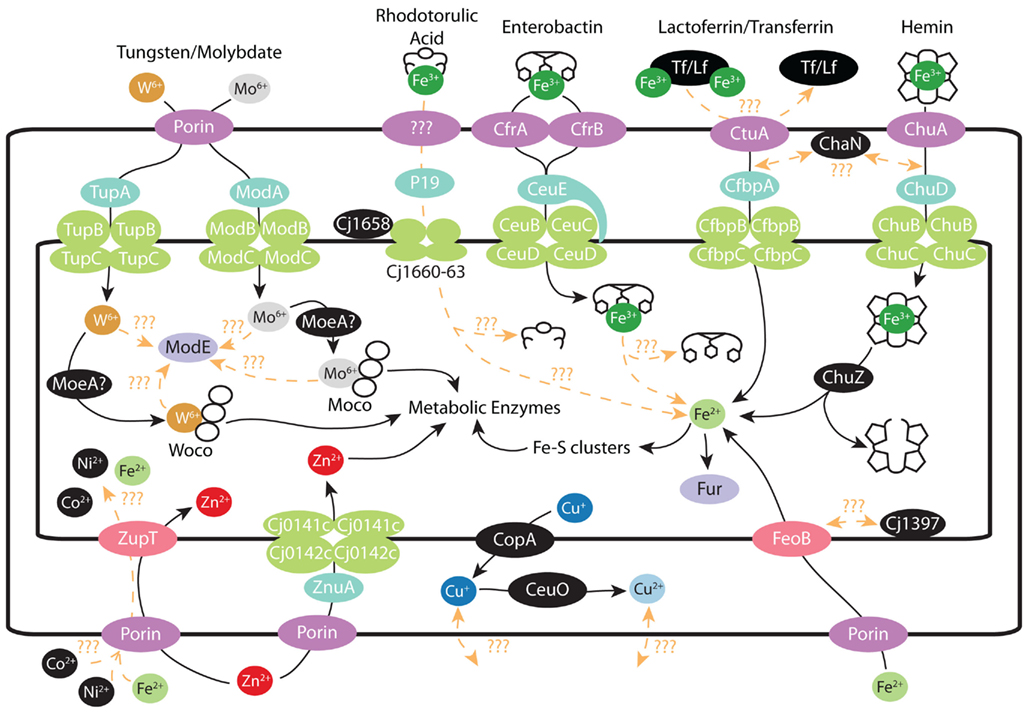
Figure 2. Metal transport in C. jejuni: schematic of various metal transport systems present in C. jejuni. Where possible, proteins are colored by their function. Outer membrane proteins are denoted by dark purple, periplasmic proteins by turquoise, ABC transporters by green, periplasmic transporters by pink, and transcriptional regulators by light purple. Iron: ferric–rhodotorulic acid is imported into the periplasm though an unidentified outer membrane transporter and subsequent interactions with various proteins result in iron release and import into the cytosol. Ferric–enterobactin is imported through either CfrA/CfrB and enters the cytosol via the CeuBCDE system. The ferric–enterobactin is subsequently hydrolyzed by an unidentified protein. Lactoferrin/Transferrin iron uptake is accomplished though CtuA and released iron is then transported thought the CfbpABC system into the cytosol. Heme is imported through the outer membrane by ChuA and transported into the cytosol via the ChuBCD system. Iron release is accomplished by ChuZ hydrolyzing the porphyrin ring. Iron imported as ferrous ions are transported by the high affinity FeoB transporter. Ferrous ions may also be non-specifically imported though the general cation imported ZupT as seen in E. coli. Iron levels are regulated by the transcriptional regulator Fur by its binding of ferrous ions. Tungsten/Molybdate: tungsten/molybdate presumably cross the outer membrane via porins and are specifically taken up by the TupABC and ModABC transport systems respectively. Tungsten/molybdate are presumably incorporated into Woco and Moco cofactors by separate MoeA enzymes for insertion into metabolic enzymes. ModE senses either free molybdate/tungstate oxyanions or Moco/Woco complexes though unidentified partners. Zinc/Copper/Others: zinc crosses the outer membrane and is taken up into the cytosol by either high affinity (ZnuA) or low affinity (ZupT) transporters. Excess cuprous ions are effluxed into the periplasm by CopA, with concomitant oxidization by CeuO into cupric ions. Other metals may cross the outer membrane through porins and imported by ZupT or there may be unidentified specific transport systems for these metals.
Ferric–rhodotorulic acid is imported into the periplasm though an unidentified outer membrane transporter. Subsequent interactions with P19 (Cj1659), Cj1658 and Cj1660–1663, and other unidentified proteins are required for import into the cytosol and release of the bound iron. It is unclear whether the entire ferric–rhodorulic acid complex enters the cytosol or if iron release occurs in the periplasm. Ferric–enterobactin is imported through either CfrA(Cj755)/CfrB(Cj0444/Cjj81176_0471) and enters the cytosol via the CeuBCDE (Cj1352–55) system (note that CeuE is a lipoprotein). The ferric–enterobactin is subsequently hydrolyzed by an unidentified protein. To note, the ferric–enterobactin complex could also be hydrolyzed within the periplasm and released iron transported through other means. Lactoferrin/Transferrin iron uptake is accomplished though CtuA (Cj0177) via an unidentified mechanism. Released iron is then transported thought the CfbpABC (Cj0173c–Cj0175c) system into the cytosol. Heme is imported through the outer membrane by ChuA (Cj1614) and transported into the cytosol via the ChuBCD (Cj1615–1617) system. ChuZ (Cj1613c) hydrolyzes the porphyrin ring to release the bound iron. It is currently unclear whether ChaN (Cj0177) functions in transferrin/lactoferrin or heme transport. Previous work has shown that ChaN forms dimers that bind heme, but the chaN gene is located directly upstream the lactoferrin/transferrin transporter (CtuA; Chan et al., 2006). Iron levels are regulated by the transcriptional regulator Fur by its binding of ferrous ions. There is also one C. jejuni strain that may be able to uptake ferrichrome as well, as it contains homologs of the E. coli FhuABD system (Galindo et al., 2001). However this pathway is not present in any of the sequenced C. jejuni strains (Miller et al., 2009).
The presence of ferric–enterobactin import in C. jejuni is particularly intriguing. Enterobactin is the prototypical bacterial siderophore that is produced to scavenge iron from host proteins such as lactoferrin or transferrin. C. jejuni does not produce enterobactin and instead takes advantage of enterobactin production from the host microbiota (Miller et al., 2009; Butcher et al., 2010). CfrA and CfrB are the two characterized enterobactin transporters in Campylobacter (Palyada et al., 2004; Carswell et al., 2008; Zeng et al., 2009; Xu et al., 2010). Curiously C. coli species harbor both proteins; while C. jejuni species only express one protein of the pair, with the other existing as a pseudogene (Xu et al., 2010). CfrB appears to be the dominant enterobactin transporter in C. coli with CfrA playing a relatively minor role. The situation in C. jejuni is relatively more complex. Many C. jejuni strains that express CfrB are not able to utilize ferric–enterobactin as a sole iron source; however CfrB is an important colonization factor for those strains that do possess a functional ferric–enterobactin pathway (Xu et al., 2010). Almost all CfrA expressing C. jejuni strains can utilize ferric–enterobactin as a sole iron source and CfrA has been demonstrated to be essential for chick colonization in these strains (Palyada et al., 2004; Xu et al., 2010). It is still unclear why Campylobacter genomes would contain two different transporters for the same iron source. It is also unknown why some strains that contain either CfrA or CfrB are still unable to utilize enterobactin as a sole iron source, when they appear to contain all the other necessary components (Xu et al., 2010).
Most deletion mutants in iron acquisition pathways show colonization defects. Studies have demonstrated that the deletion of the transporters responsible for the ferrous ion (FeoB; Naikare et al., 2006), lactoferrin/transferrin (CtuA; Miller et al., 2008), and enterobactin (CfrA/CfrB; Palyada et al., 2004; Xu et al., 2010) transporters resulted in substantial colonization deficiencies in the chicks (Palyada et al., 2004; Stintzi et al., 2005; Naikare et al., 2006). The feoB and ctuA mutants are also defective in colonizing pathogenic animal models, as demonstrated using either the colostrum-deprived piglet or the rabbit ileal loop model respectively (Stintzi et al., 2005; Naikare et al., 2006). Curiously, the abrogation of the heme (ChuA; Ridley et al., 2006) or rhodotorulic acid (P19/Cj1658; Stintzi et al., 2008) iron uptake pathways had no effect on C. jejuni chick colonization, although these pathways might be important for colonization in other hosts or environments (Butcher et al., 2010).
Molybdate and Tungstate
Campylobacter jejuni is fairly unique amongst bacteria in that it possesses both molybdenum (Moco) and tungsten (Woco) cofactored proteins. C. jejuni formate dehydrogenase, a key enzyme for energy metabolism using formate as an electron donor, has been shown to require Woco for proper function, while nitrate reductase, sulfite oxidase, and SN oxide reductase (Cj0264c) require Mocos for their activity (Smart et al., 2009; Taveirne et al., 2009). This peculiar metabolic need likely explains the unusual presence in C. jejuni of specific high affinity ABC transporters for both metals (Figure 2). The ModABC (Cj0300c–Cj0303c) and TupABC (Cj1538c–Cj1540) transporters import either molybdate or tungstate respectively across the inner membrane in an ATP dependant manner, but both are regulated together by a single ModE-like regulatory protein (Cj1507c; Taveirne et al., 2009). Interestingly, the C. jejuni ModE protein represses the mod operon in the presence of excess molybdate or tungstate while the tup operon is only repressed in response to tungsten. Moreover the C. jejuni ModE protein does not appear to contain a metal binding domain and instead contains a coiled–coiled domain that is indicative of protein–protein interactions (Taveirne et al., 2009). It is currently unclear what other factor(s) are required to sense excess intracellular molybdate/tungsten in C. jejuni. One possibility is that C. jejuni contains alternate molybdenum/tungsten sensors that interact with ModE to regulate each operon as needed. It is also possible that ModE senses the concentration of molybdenum and tungsten indirectly though the binding of Moco or Woco respectively.
The presence of both molybdate and tungsten requiring enzymes in C. jejuni also creates additional metabolic challenges. Most bacteria, such as E. coli, only use Moco cofactored enzymes and these enzymes are normally inactivated if tungsten is misincorporated during cofactor synthesis (Kletzin and Adams, 1996). This is not normally a problem, as these bacteria typically do not have specific high affinity transporters for tungstate. However, C. jejuni has specific high affinity transporters for both molybdate and tungstate and also possesses both Moco and Woco dependent enzymes. Thus, C. jejuni must contain proteins that can distinguish between molybdate and tungsten for the proper synthesis of these distinct cofactors. C. jejuni must also be able to distinguish between the produced Moco and Woco cofactors to correctly insert each cofactor into the correct effector proteins. In fact, Smart et al. (2009) have proposed that these needs are the reason why C. jejuni genomes contain two versions of the molydopterin synthesis protein moeA (Cj0857c/Cj1519). It remains to be demonstrated if each moeA protein is either Moco or Woco specific and how effector proteins ensure that they receive the proper cofactor.
Campylobacter jejuni mutants deficient in either molybdate or tungstate transport have not yet been tested for their ability to colonize in vivo models. However, deletion of tungstate acquisition should result in reduced colonization potential, as deletion of the tungsten dependent formate dehydrogenase in C. jejuni resulted in chick colonization defects (Weerakoon et al., 2009).
Copper/Zinc
There is currently very little known about the mechanisms surrounding copper and zinc homeostasis in C. jejuni. Copper chelation results in slowed C. jejuni growth and this growth inhibition can be partially relieved by deleting non-essential copper containing proteins such as the p19 iron acquisition protein (Chan et al., 2010). C. jejuni does not have any annotated homologs of the copper regulators CueR or CusR but does contain CopA (Cj1161) and CeuO (Cj1516) homologs (Hall et al., 2008; Figure 2). CopA proteins are transmembrane ATPases that are involved in pumping excess cytosolic copper to the periplasmic space (Osman and Cavet, 2008). Cuprous ions in the periplasmic space are oxidized to the less toxic cupric form by CeuO (Osman and Cavet, 2008). Deleting C. jejuni copA results in a severe sensitivity toward excess copper levels (Hall et al., 2008). CeuO deletion also results in a copper sensitivity phenotype (Hall et al., 2008). CopA and CeuO probably function independently from each other since a double deletion mutant in both genes is more sensitive to copper levels then either single deletion mutant (Hall et al., 2008). It is currently unknown whether C. jejuni contains specific copper transporters for copper import or export from the cell.
The zinc homeostasis system remains fairly uncharacterized in C. jejuni. There are no obvious candidates for the regulation of zinc uptake or efflux despite the almost universal presence of these proteins in other bacteria (Hantke, 2001). C. jejuni does possess a homolog of the high efficiency E. coli ZnuABC zinc uptake system (Cj0141c–Cj0143c) that is also present in many bacteria (Hantke, 2001). Recent work by Davis et al. (2009) has demonstrated that this system is functional in C. jejuni and is required for both survival in low zinc environments and for successful chick colonization. C. jejuni also contains a homolog of the low affinity zinc transporter ZupT, however, there has not as of yet been any studies conducted to demonstrate how ZupT may function in C. jejuni. As previously mentioned, in E. coli, this protein can also transport various other divalent cations including Fe2+, Co2+, and Mn2+ (Grass et al., 2005).
Other Metals
Bacteria such as C. jejuni also make use of several additional metals for either enzymatic or structural purposes, such as cobalt and nickel. While there are no obvious transporters for these metals in the C. jejuni genome, there are several examples where these metals are required for proper protein function. For instance, the C. jejuni NeuB3 (Cj1317) has been shown to rely on cobalt for full enzymatic activity (Chou et al., 2005). Another example is the C. jejuni hydrogenase complex (Cj1263c–1265c), which is predicted to be nickel dependent (Parkhill et al., 2000; Weerakoon et al., 2009). The acquisition of these metals may be accomplished through general cation transporters (e.g., ZupT) or accomplished through uncharacterized metal specific transporters.
Conclusion
While the C. jejuni metabolic networks and nutrient acquisition systems have remained to a significant extent a mystery, even after the sequencing of the genome, researchers are gradually piecing together a better picture of how this organism functions. Once thought to be completely asaccharolytic, we now know that it possesses a L-fucose metabolic pathway. Its usage of amino acids and CAC intermediates are coming into focus, and we are gaining a better understanding of its complex metal acquisition pathways, especially those involving iron, molybdate, and tungsten. Although a great deal still remains obscure, the understanding of these pathways is contributing substantially to our understanding of this common gastrointestinal pathogen.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research in Dr. Stintzi’s laboratory is supported by the Canadian Institutes of Health Research (CIHR).
References
Baldoma, L., and Aguilar, J. (1988). Metabolism of L-fucose and L-rhamnose in Escherichia coli: aerobic-anaerobic regulation of L-lactaldehyde dissimilation. J. Bacteriol. 170, 416–421.
Barnes, I. H., Bagnall, M. C., Browning, D. D., Thompson, S. A., Manning, G., and Newell, D. G. (2007). Gamma-glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb. Pathog. 43, 198–207.
Belenguer, A., Holtrop, G., Duncan, S. H., Anderson, S. E., Calder, A. G., Flint, H. J., and Lobley, G. E. (2011). Rates of production and utilization of lactate by microbial communities from the human colon. FEMS Microbiol. Ecol. 77, 107–119.
Butcher, J., Flint, A., Stahl, M., and Stintzi, A. (2010). “Campylobacter Fur and PerR regulons,” in Iron Uptake and Homeostasis in Microorganisms, eds P. Cornelis and S. C. Andrews (Norfolk: Caister Academic Press), 167–202.
Carswell, C. L., Rigden, M. D., and Baenziger, J. E. (2008). Expression, purification, and structural characterization of CfrA, a putative iron transporter from Campylobacter jejuni. J. Bacteriol. 190, 5650–5662.
Chan, A. C., Doukov, T. I., Scofield, M., Tom-Yew, S. A., Ramin, A. B., Mackichan, J. K., Gaynor, E. C., and Murphy, M. E. (2010). Structure and function of P19, a high-affinity iron transporter of the human pathogen Campylobacter jejuni. J. Mol. Biol. 401, 590–604.
Chan, A. C., Lelj-Garolla, B. I., Rosell, F., Pedersen, K. A., Mauk, A. G., and Murphy, M. E. (2006). Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J. Mol. Biol. 362, 1108–1119.
Chou, W. K., Dick, S., Wakarchuk, W. W., and Tanner, M. E. (2005). Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J. Biol. Chem. 280, 35922–35928.
Coyne, M. J., Reinap, B., Lee, M. M., and Comstock, L. E. (2005). Human symbionts use a host-like pathway for surface fucosylation. Science 307, 1778–1781.
Dang, S., Sun, L., Huang, Y., Lu, F., Liu, Y., Gong, H., Wang, J., and Yan, N. (2010). Structure of a fucose transporter in an outward-open conformation. Nature 467, 734–738.
Davis, L. M., Kakuda, T., and DiRita, V. J. (2009). A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J. Bacteriol. 191, 1631–1640.
del Rocio Leon-Kempis, M., Guccione, E., Mulholland, F., Williamson, M. P., and Kelly, D. J. (2006). The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60, 1262–1275.
Duncan, S. H., Holtrop, G., Lobley, G. E., Calder, A. G., Stewart, C. S., and Flint, H. J. (2004). Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 91, 915–923.
Fauchere, J. L., Kervella, M., Rosenau, A., Mohanna, K., and Veron, M. (1989). Adhesion to HeLa cells of Campylobacter jejuni and C. coli outer membrane components. Res. Microbiol. 140, 379–392.
Flint, D. H., Smyk-Randall, E., Tuminello, J. F., Draczynska-Lusiak, B., and Brown, O. R. (1993). The inactivation of dihydroxy-acid dehydratase in Escherichia coli treated with hyperbaric oxygen occurs because of the destruction of its Fe-S cluster, but the enzyme remains in the cell in a form that can be reactivated. J. Biol. Chem. 268, 25547–25552.
Galindo, M. A., Day, W. A., Raphael, B. H., and Joens, L. A. (2001). Cloning and characterization of a Campylobacter jejuni iron-uptake operon. Curr. Microbiol. 42, 139–143.
Grass, G., Franke, S., Taudte, N., Nies, D. H., Kucharski, L. M., Maguire, M. E., and Rensing, C. (2005). The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J. Bacteriol. 187, 1604–1611.
Guccione, E., Hitchcock, A., Hall, S. J., Mulholland, F., Shearer, N., van Vliet, A. H., and Kelly, D. J. (2010). Reduction of fumarate, mesaconate and crotonate by Mfr, a novel oxygen-regulated periplasmic reductase in Campylobacter jejuni. Environ. Microbiol. 12, 576–591.
Guccione, E., Leon-Kempis Mdel, R., Pearson, B. M., Hitchin, E., Mulholland, F., van Diemen, P. M., Stevens, M. P., and Kelly, D. J. (2008). Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69, 77–93.
Hall, S. J., Hitchcock, A., Butler, C. S., and Kelly, D. J. (2008). A multicopper oxidase (Cj1516) and a CopA homologue (Cj1161) are major components of the copper homeostasis system of Campylobacter jejuni. J. Bacteriol. 190, 8075–8085.
Hofreuter, D., Novik, V., and Galan, J. E. (2008). Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4, 425–433.
Hofreuter, D., Tsai, J., Watson, R. O., Novik, V., Altman, B., Benitez, M., Clark, C., Perbost, C., Jarvie, T., Du, L., and Galán, J. E. (2006). Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74, 4694–4707.
Kletzin, A., and Adams, M. W. (1996). Tungsten in biological systems. FEMS Microbiol. Rev. 18, 5–63.
Krishnan, N., Doster, A. R., Duhamel, G. E., and Becker, D. F. (2008). Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect. Immun. 76, 3037–3044.
Lee, M. D., and Newell, D. G. (2006). Campylobacter in poultry: filling an ecological niche. Avian Dis. 50, 1–9.
Lin, A. E., Krastel, K., Hobb, R. I., Thompson, S. A., Cvitkovitch, D. G., and Gaynor, E. C. (2009). Atypical roles for Campylobacter jejuni amino acid ATP binding cassette transporter components PaqP and PaqQ in bacterial stress tolerance and pathogen-host cell dynamics. Infect. Immun. 77, 4912–4924.
Line, J. E., Hiett, K. L., Guard-Bouldin, J., and Seal, B. S. (2010). Differential carbon source utilization by Campylobacter jejuni 11168 in response to growth temperature variation. J. Microbiol. Methods 80, 198–202.
Louis, P., Young, P., Holtrop, G., and Flint, H. J. (2010). Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314.
Mendz, G. L., Ball, G. E., and Meek, D. J. (1997). Pyruvate metabolism in Campylobacter spp. Biochim. Biophys. Acta 1334, 291–302.
Miller, C. E., Rock, J. D., Ridley, K. A., Williams, P. H., and Ketley, J. M. (2008). Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J. Bacteriol. 190, 1900–1911.
Miller, C. E., Williams, P. H., and Ketley, J. M. (2009). Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology 155, 3157–3165.
Muraoka, W. T., and Zhang, Q. (2010). Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 193, 1065–1075.
Naikare, H., Palyada, K., Panciera, R., Marlow, D., and Stintzi, A. (2006). Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74, 5433–5444.
Nakajima, K., Inatsu, S., Mizote, T., Nagata, Y., Aoyama, K., Fukuda, Y., and Nagata, K. (2008). Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed. Res. 29, 9–18.
Osman, D., and Cavet, J. S. (2008). Copper homeostasis in bacteria. Adv. Appl. Microbiol. 65, 217–247.
Palyada, K., Threadgill, D., and Stintzi, A. (2004). Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186, 4714–4729.
Parker, C. T., Miller, W. G., Horn, S. T., and Lastovica, A. J. (2007). Common genomic features of Campylobacter jejuni subsp. doylei strains distinguish them from C. jejuni subsp. jejuni. BMC Microbiol. 7, 50. doi:10.1186/1471-2180-7-50
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., Chillingworth, T., Davies, R. M., Feltwell, T., Holroyd, S., Jagels, K., Karlyshev, A. V., Moule, S., Pallen, M. J., Penn, C. W., Quail, M. A., Rajandream, M. A., Rutherford, K. M., van Vliet, A. H., Whitehead, S., and Barrell, B. G. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668.
Parsons, C. M. (1984). Influence of caecectomy and source of dietary fibre or starch on excretion of endogenous amino acids by laying hens. Br. J. Nutr. 51, 541–548.
Pei, Z., Burucoa, C., Grignon, B., Baqar, S., Huang, X. Z., Kopecko, D. J., Bourgeois, A. L., Fauchere, J. L., and Blaser, M. J. (1998). Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66, 938–943.
Ridley, K. A., Rock, J. D., Li, Y., and Ketley, J. M. (2006). Heme utilization in Campylobacter jejuni. J. Bacteriol. 188, 7862–7875.
Robinson, D. A. (1981). Infective dose of Campylobacter jejuni in milk. Br. Med. J. (Clin. Res. Ed.) 282, 1584.
Smart, J. P., Cliff, M. J., and Kelly, D. J. (2009). A role for tungsten in the biology of Campylobacter jejuni: tungstate stimulates formate dehydrogenase activity and is transported via an ultra-high affinity ABC system distinct from the molybdate transporter. Mol. Microbiol. 74, 742–757.
Stahl, M., Friis, L. M., Nothaft, H., Liu, X., Li, J., Szymanski, C. M., and Stintzi, A. (2011). L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U.S.A. 108, 7194–7199.
Stintzi, A., Marlow, D., Palyada, K., Naikare, H., Panciera, R., Whitworth, L., and Clarke, C. (2005). Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73, 1797–1810.
Stintzi, A., van Vliet Arnoud, H. M., and Ketley, J. M. (2008). “Iron metabolism, transport, and regulation,” in Campylobacter, eds I. Nachamkin, C. M. Szymanski, and M. J. Blaser (Washington, DC: ASM Press), 591.
Taveirne, M. E., Sikes, M. L., and Olson, J. W. (2009). Molybdenum and tungsten in Campylobacter jejuni: their physiological role and identification of separate transporters regulated by a single ModE-like protein. Mol. Microbiol. 74, 758–771.
Thomas, M. T., Shepherd, M., Poole, R. K., van Vliet, A. H., Kelly, D. J., and Pearson, B. M. (2011). Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate. Environ. Microbiol. 13, 48–61.
van Vliet, A. H., Ketley, J. M., Park, S. F., and Penn, C. W. (2002). The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26, 173–186.
Velayudhan, J., Jones, M. A., Barrow, P. A., and Kelly, D. J. (2004). L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72, 260–268.
Velayudhan, J., and Kelly, D. J. (2002). Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology 148, 685–694.
Weerakoon, D. R., Borden, N. J., Goodson, C. M., Grimes, J., and Olson, J. W. (2009). The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 47, 8–15.
Weingarten, R. A., Taveirne, M. E., and Olson, J. W. (2009). The dualcfunctioning fumarate reductase is the sole succinate:quinone reductase in Campylobacter jejuni and is required for full host colonization. J. Bacteriol. 191, 5293–5300.
Wright, J. A., Grant, A. J., Hurd, D., Harrison, M., Guccione, E. J., Kelly, D. J., and Maskell, D. J. (2009). Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155, 80–94.
Xu, F., Zeng, X., Haigh, R. D., Ketley, J. M., and Lin, J. (2010). Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J. Bacteriol. 192, 4425–4435.
Yew, W. S., Fedorov, A. A., Fedorov, E. V., Rakus, J. F., Pierce, R. W., Almo, S. C., and Gerlt, J. A. (2006). Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry 45, 14582–14597.
Zeng, X., Xu, F., and Lin, J. (2009). Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect. Immun. 77, 5437–5448.
Keywords: Campylobacter jejuni, metabolism, metal transport, nutrient acquisition
Citation: Stahl M, Butcher J and Stintzi A (2012) Nutrient acquisition and metabolism by Campylobacter jejuni. Front. Cell. Inf. Microbio. 2:5. doi: 10.3389/fcimb.2012.00005
Received: 29 November 2011;
Accepted: 21 January 2012;
Published online: 07 February 2012.
Edited by:
D. Scott Merrell, Uniformed Services University of the Health Sciences, USAReviewed by:
Erin Gaynor, University of British Columbia, CanadaArnoud H. M. Van Vliet, Institute of Food Research, UK
Copyright: © 2012 Stahl, Butcher and Stintzi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Alain Stintzi, Ottawa Institute of Systems Biology, Department of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, 451 Smyth Road, Ottawa, ON, Canada K1H 8M5. e-mail: astintzi@uottawa.ca