- Centre for Bacterial Cell Biology, Institute for Cell and Molecular Biosciences, Medical School, Newcastle University, Newcastle upon Tyne, UK
One of the most important achievements of bacteria is its capability to adapt to the changing conditions of the environment. The competition for nutrients with other microorganisms, especially in the soil, where nutritional conditions are more variable, has led bacteria to evolve a plethora of mechanisms to rapidly fine-tune the requirements of the cell. One of the essential nutrients that are normally found in low concentrations in nature is inorganic phosphate (Pi). Bacteria, as well as other organisms, have developed several systems to cope for the scarcity of this nutrient. To date, the unique mechanism responding to Pi starvation known in detail is the Pho regulon, which is normally controlled by a two component system and constitutes one of the most sensible and efficient regulatory mechanisms in bacteria. Many new members of the Pho regulon have emerged in the last years in several bacteria; however, there are still many unknown questions regarding the activation and function of the whole system. This review describes the most important findings of the last three decades in relation to Pi regulation in bacteria, including: the PHO box, the Pi signaling pathway and the Pi starvation response. The role of the Pho regulon in nutritional regulation cross-talk, secondary metabolite production, and pathogenesis is discussed in detail.
Introduction
Phosphorus, in terms of cellular content, is the fifth most important element. It is essential for several important biological processes such as the inheritance of genetic materials, energy metabolism, membrane integrity, and intracellular signaling. The availability of this nutrient has many medical, agricultural, and pharmaceutical implications. Misregulation of phosphorus can cause serious human disorders like tumor-induced osteomalacia, muscle myopathy, cardiomyopathy, neuropathy, and haemolysis, and in some cases, it can contribute to death (Bergwitz and Jüppner, 2011). In agriculture, phosphorus is required to ensure high levels of agricultural productivity (Ha and Tran, 2014). With this aim, modern agricultural practice extensively uses fertilizers, in many cases producing negative environmental effects. From an industrial point of view, phosphorous availability is also quite important. Pharmaceutical companies commercialize an impressive number of antibiotics every year and other valuable secondary metabolites such as antifungal compounds, anti-tumor agents, immuno-suppressants, and pigments (Hopwood, 2007). The production of most of these compounds is quite reduced by high amounts of phosphorous in the fermentation tanks (Martin, 2004). Therefore, companies are often forced to use low concentrations of this nutrient which results in a significant decrease of the growth yields.
The main assimilable form of phosphorous in bacteria is the orthophosphate anion (PO43-), most commonly known as inorganic phosphate (Pi). Despite the essentiality of Pi for life, this nutrient is usually found at very low concentrations in the environment, especially in soil. To adapt and survive to Pi scarcity, bacteria, as well as yeasts, plants and animals, have evolved physiological and biochemical responses aimed to acquire and save this nutrient. Whether these mechanisms in eukaryotes involve similar or different bacterial systems is unknown. This review provides a critical overview of the most important insights regarding Pi regulation in bacteria, for example antibiotic production, nutritional stress networking, or pathogenesis.
The Pho Regulon and the PHO Box
The Phosphate (Pho) regulon is a global regulatory mechanism involved in bacterial Pi management that was first characterized in Escherichia coli, and later in many other bacterial species (Wanner and Chang, 1987). The most common members activated by the Pho regulon are: extracellular enzymes capable of obtaining Pi from organic phosphates, Pi-specific transporters, and enzymes involved in storage and saving of the nutrient. The Pst Pi-specific transporter is the most conserved member of the Pho regulon in all bacteria. Other Pi transporters commonly found in bacteria, such as the low affinity Pi transporter Pit, are regulated in a variable fashion manner in the different species (Santos-Beneit et al., 2008). E. coli activates additional transporters for phosphorous-containing compounds, such as Ugp (i.e., glycerophosphodiesters uptake) and Phn (i.e., phosphonates uptake). Among the Pi scavengers, the alkaline phosphatases (PhoA), phospholipases (PhoD), glycerophosphodiester phosphodiesterases (GlpQ and UgpQ), phytases (PhyC) and 5′-nucleotidases (UshA) are the most common enzymes induced in response to Pi starvation in bacteria (Wanner and Chang, 1987). For the storage of Pi, most bacteria induce the expression of PpK, which is able to accumulate polyphosphate as a Pi reservoir and, when needed, reuse it (Ghorbel et al., 2006a). For saving nutrients, some bacteria are able to replaced teichoic acids (Pi-rich polymers found in the cell wall of Gram-positive bacteria) by teichuronic acids (Pi-free polymers). The biosynthetic genes are repressed and activated, respectively, by the Pho regulon (Liu et al., 1998).
The Pho regulon is controlled by a two-component regulatory system which comprises an inner-membrane histidine kinase sensor protein and a cytoplasmic transcriptional response regulator. These proteins have received different names in some bacteria, such as: PhoR–PhoB in E. coli (Tommassen et al., 1982), PhoR–PhoP in Bacillus subtilis (Hulett et al., 1994), PnpR–PnpS in Streptococcus pneumoniae (Novak et al., 1999), PhoR–PhoS in Corynebacterium glutamicum (Kocan et al., 2006), PhosS–PhosR in Campylobacter jejuni (Wösten et al., 2006), SphS–SphR in Synechocystis sp. (Juntarajumnong et al., 2007), and SenX3–RegX3 in Mycobacterium smegmatis (Glover et al., 2007). In all cases, upon Pi scarcity, the response regulator is phosphorylated on an aspartic residue by the sensor kinase. The phosphorylated response regulator is able to bind to specific sequences on the DNA and activate or repress the transcription of genes. These specific sequences, known as PHO boxes, were first characterized in E. coli (Makino et al., 1988). Initially, the PHO boxes were defined as 18-nucleotide sequences comprising two 7-nucleotide direct repeat units separated by four non-conserved nucleotides. However, after the crystallographic studies by Blanco et al. (2002), the PHO box was defined as the sum of two 11-nucleotide direct repeat units composed of seven well conserved and four less conserved nucleotides in each. Similar features have also been observed in many other bacteria, among others: B. subtilis, Sinorhizobium meliloti, C. glutamicum, and Streptomyces coelicolor (Hulett, 1996; Yuan et al., 2006; Schaaf and Bott, 2007; Sola-Landa et al., 2008). The consensus sequence of the PHO box varies from one species to another and depends on the specific contacts that the amino acids of the response regulator are able to eject with the bases of the DNA (Blanco et al., 2002). For instance, the most conserved motif of the PHO box in E. coli is CTGTCAT (Yoshida et al., 2012) while in S. coelicolor is GTTCACC (Sola-Landa et al., 2008); this is not strange since the two microorganisms have a distinct GC content in its genomes. Nonetheless, the PHO boxes of diverse promoters in a specific bacterium contain distinct DNA sequences, which results in different protein-DNA binding affinities. This has been well documented in S. coelicolor, in which a high degree of variation in the sequence conservation, number, and organization of the different direct repeat units that form the PHO boxes has been noticed (Martín et al., 2012). According to this variation, the Pho operators in this bacterium have been classified as simple or complex operators. It is important to note that several experimentally confirmed PHO boxes are found in genes that lack an obvious Pi regulation (at least under a given condition) and that, on the contrary, clear Pho regulon members lack any conserved PHO boxes (Allenby et al., 2012; Martín et al., 2012). This might indicate the existence of additional features in the PHO boxes and/or extra regulatory proteins or ligands involved in the control of the PHO regulon. In this sense, the interaction of the response regulator with the RNA polymerase sigma subunit has been well reported for triggering transcription initiation in both E. coli and B. subtilis. In E. coli, the σ70 subunit is involved in this interaction, while at least two different sigma factors (σA and σE) may be involved in B. subtilis (Makino et al., 1993; Paul et al., 2004). Recently, Blanco et al. (2011) have provided crystallographic evidence that PhoB directly contacts the C-terminal region of σ70 to accomplish RNA polymerase recruitment in E. coli.
The features for dimerization and oligomerization of PhoB (E. coli) and PhoP (B. subtilis) on the DNA differ. In E. coli, PhoB is already a dimer before binding to the DNA, but it has to be phosphorylated in order to become active (Bachhawat et al., 2005). The oligomerization of the active dimers on the DNA is achieved in a hierarchical and cooperative manner and is favored by protein–protein interactions (Blanco et al., 2012). PhoB presents two structural domains connected by a flexible linker: (i) the N-terminal receiver domain (RD) containing the conserved aspartate (Asp53) to which a phosphoryl group is transferred from the histidine (His215) of PhoR and (ii) the C-terminal effector domain (ED), which presents a winged-helix fold with the specific DNA binding and transactivation properties (Solá et al., 1999). Phosphorylation of the RD is needed to switch on the ED, although if the RD is removed, the ED can constitutively bind to the DNA (Ellison and McCleary, 2000). Different structural analyses have determined that the RD dimerises with twofold rotational symmetry (i.e., head-to-head). This orientation contrasts with the binding of the ED to the direct repeats units of the PHO boxes in a tandem head-to-tail orientation (Blanco et al., 2002). Bachhawat et al. (2005) have proposed a model which implies an obligatory rotation of the flexible linker connecting the RD and ED domains when they are in their active state; avoiding the problem of rotation (see Figure 1). In B. subtilis, PhoP dimerises asymmetrically by using two distinct faces of the RD, being able to form a great variety of multimers on the DNA (Chen et al., 2003).
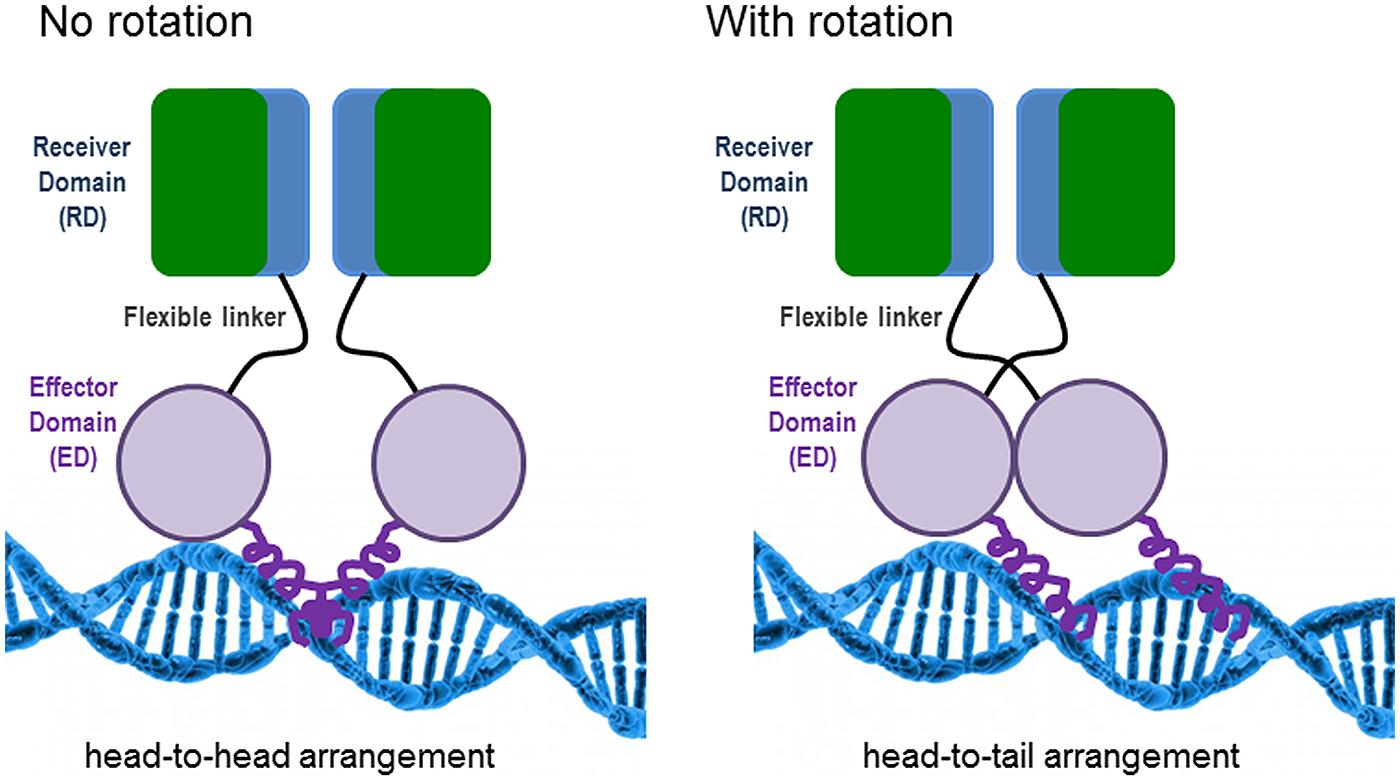
FIGURE 1. Model of PhoB binding to the DNA in Escherichia coli according to the work of Bachhawat et al. (2005). The non-rotational arrangement would provide an inactive state by positioning the effector domains (EDs) in opposite directions, incompatible with tandem DNA binding. In the rotational arrangement, the receiver domains (RDs) would form a twofold symmetric dimer, while the EDs would bind to DNA with tandem symmetry.
Pi Sensing and Signal Transduction Pathway
Inorganic phosphate depletion in the medium has been shown to be essential for activation of the Pho regulon in bacteria; differing with other microorganisms such as Saccharomyces cerevisiae, in which the intracellular Pi concentration seems to account for this activation (Auesukaree et al., 2004). The complete signal transduction system that senses and transmits the Pi scarcity signal to the response regulator has not been yet completely elucidated and seems to vary from one organism to another.
In E. coli, the Pi-sensing pathway requires, apart from the two-component system PhoR–PhoB, five additional proteins. Four of those are components of the Pi-specific transporter Pst (PstSCAB) and the other is the metal binding protein, PhoU. Under Pi limitation, PhoB is activated by PhoR acting as a kinase, but under Pi-replete conditions, PhoB activation is interrupted by PhoR acting as a phosphatase (Carmany et al., 2003). PhoU is required for PhoB dephosphorylation under Pi-rich conditions in an as yet unknown manner. When phoU is mutated or deleted, PhoR behaves as a constitutive PhoB kinase, leading to high expression of the Pho regulon genes (Steed and Wanner, 1993; Rice et al., 2009). PhoU is involved not only in control of the autokinase activity of PhoR, but also in the control of the Pst system in order to avoid an uncontrolled Pi uptake that could be toxic for the cell (Surin et al., 1986; Muda et al., 1992; Carmany et al., 2003; Gardner et al., 2014). Whether it is PstSCAB, PhoR, PhoU, or another player the component that senses the Pi scarcity is still unknown. Since PhoR does not contain a significant periplasmic sensory domain, it makes sense to assume that the Per-Arnt-Sim (PAS) domain of the protein senses a cytoplasmic signal; although the nature of the signal is not known.
Curiously, although phoU is found in many bacterial genomes, B. subtilis lacks this gene. The Pi signal transduction network in this bacterium includes a positive feedback loop between the PhoP–PhoR and ResD–ResE two-component systems (Schau et al., 2004). Alternative players such as the Spo0B–Spo0A two-component system, the AbrB and ScoC transition state regulators and an intermediate of the teichoic acid synthesis pathway are also involved in this network (Hulett, 1996; Sun et al., 1996; Kaushal et al., 2010; Botella et al., 2014). ResD activates genes involved in both aerobic and anaerobic respiration, including two terminal oxidase genes. ResD is required for 80% of the wild-type B. subtilis Pho response. This transcriptional regulator does not bind to the phoPR operon and seems to eject its control through the expression of terminal oxidases (such as ctaA or cytochrome bd) that are able to regulate the autophosphorylation activity of PhoR, although the mechanism of this control is not known. PhoP induces the expression of resDE, indicating a feedback loop between these two component systems (Birkey et al., 1998).
Disruption of the abrB transition state regulator causes a 20% reduction of the wild-type B. subtilis Pho response. Actually, a resD–abrB double mutant shows no Pho induction and resembles the phenotype of a phoPR mutant strain (Hulett et al., 1994). The other transition state regulator involved in the network (ScoC), contrary to AbrB, produces a negative effect on the expression of phoPR (Kaushal et al., 2010). This negative effect is accumulative with the one ejected by another B. subtilis regulator, CcpA, which is involved in carbon metabolism (Puri-Taneja et al., 2006; see below). Spo0B–Spo0A attenuates the Pho response by negative regulation of both ResE–ResD and AbrB pathways (Strauch et al., 1990; Prágai et al., 2004), connecting the control of B. subtilis sporulation (i.e., Spo0B–Spo0A) with the Pi response. Recently, Botella et al. (2014) have shown new insights in the activation of the B. subtilis Pho system. These authors have reported that the PhoR autokinase activity is inhibited by an intracellular intermediate of teichoic acid biosynthesis which is sensed through the cytoplasmic PAS domain of the protein. These two mechanisms of Pi signal transduction (i.e., E. coli and B. subtilis) may not be fully conserved in other bacteria. For example, no constitutive induction of the Pho regulon has been displayed in pst and phoU mutants of several Streptomyces species (Díaz et al., 2005; Ghorbel et al., 2006b). On the other hand, as mentioned above, several cross-talks among two-component systems have been reported in E. coli and B. subtilis. Actually, at least seven different kinases are able to phosphorylate PhoB in E. coli (Amemura et al., 1990; Fisher et al., 1995; Zhou et al., 2005). This promiscuity of the Pho system is not observed in some bacteria, for example S. coelicolor, where PhoR appears to be the only specific sensor kinase able to phosphorylate PhoP (Fernández-Martínez et al., 2012).
Involvement of Pi Regulation in Secondary Metabolite Production
The synthesis of many classes of secondary metabolites (i.e., macrolides, tetracyclines, aminoglycosides, etc.) is negatively regulated by high Pi concentrations in the culture media (reviewed in Martin, 2004). Pi control has been profoundly studied in species of the Streptomyces genus, which provides the source of most of the antibiotic compounds in clinical use today (Hopwood, 2007). Most of these studies were performed a long time ago and report data of the overall Pi concentration effect on the control of antibiotic production, but surprisingly the molecular mechanism of this control remains still unclear. Works showing absent or retarded antibiotic production when high Pi concentrations are added to the culture medium have been performed at least in the following Streptomycetes: S. griseus (Martin and Demain, 1976), S. clavuligerus (Aharonowitz and Demain, 1977), S. fradiae (Vu-Trong et al., 1981), S. noursei (Müller et al., 1984); S. rosa (Masuma et al., 1986), S. coelicolor (Doull and Vining, 1990), S. tendae (Hege-Treskatis et al., 1992), S. acrimycini (Asturias et al., 1994), S. hygroscopicus (Cheng et al., 1995), S. rimosus (McDowall et al., 1999), S. lividans (Chouayekh and Virolle, 2002), S. venezuelae (Jakeman et al., 2006), S. natalensis (Mendes et al., 2007), S. cacaoi (Tenconi et al., 2012), and S. tsukubaensis (Martínez-Castro et al., 2013). Contrary to the large amount of studies describing the effect of the Pi concentration on antibiotic production, very few exist about the implication of the Pho regulon in this process. The effect of phoP deletion in the control of antibiotic production has been determined so far in only three bacteria: S. lividans (Sola-Landa et al., 2003), S. natalensis (Mendes et al., 2007), and S. coelicolor (Santos-Beneit et al., 2009a; Fernández-Martínez et al., 2012; Thomas et al., 2012). Sola-Landa et al. (2003) reported for the first time the involvement of the PhoR–PhoP system in the control of secondary metabolite production. These authors showed that synthesis of the antibiotics actinorhodin and undecylprodigiosin is greatly enhanced in both ΔphoP and ΔphoRP S. lividans mutants in R5 media (either solid or liquid). A similar result is obtained in R5 medium for the same antibiotics in S. coelicolor ΔphoP, a close relative of S. lividans (Fernández-Martínez et al., 2012), or in the pimaricin producer, S. natalensis, using another complex medium, NBG (Mendes et al., 2007). However, the effect of PhoR–PhoP on antibiotic production is culture media-dependent. For example, actinorhodin and undecylprodigiosin productions are repressed in a S. coelicolor ΔphoP mutant when using, instead of complex media, defined starch-containing media (Santos-Beneit et al., 2009a; Fernández-Martínez et al., 2012; Thomas et al., 2012).
The regulatory mechanism of PhoR–PhoP on antibiotic control is unknown. Only four antibiotic regulatory genes (afsS, atrA, cdaR, and scbR) have been described as members of the Pho regulon in a chromatin-immunoprecipitation-on-microarray analysis of S. coelicolor wild-type and ΔphoP mutant cultures using purified PhoP protein (Allenby et al., 2012). Unfortunately, detailed studies on this control only exist in the S. coelicolor afsS gene (Santos-Beneit et al., 2009a, 2011). AfsS is part of a signal transduction system which includes additional components, such as AfsR, AfsK, AfsL, PkaG, and KbpA (see Figure 2). Upon sensing particular signals, such as the accumulation of S-adenosyl-L-methionine (SAM), AfsR is phosphorylated by the serine/threonine kinase AfsK (Matsumoto et al., 1994; Jin et al., 2011). The KbpA protein is able to inhibit AfsK autophosphorylation (Umeyama et al., 2002). Two additional serine/threonine kinases (AfsL and PkaG) can also phosphorylate AfsR (Sawai et al., 2004), which suggests that AfsR integrates multiple signals. The phosphorylated active form of AfsR binds the -35 region of the afsS promoter and activates its transcription (Lee et al., 2002; Tanaka et al., 2007). The fate of the small protein AfsS is not known, but appears to interact with the RNA-polymerase activating the transcription of the pathway specific antibiotic regulatory genes. PhoP competes for binding to the afsS promoter region with AfsR (Santos-Beneit et al., 2009a). Both regulators behave as competitive activators on afsS, with each regulator being able to activate the transcription of the gene when acting separately, but acting as competitive proteins when occurring together (Santos-Beneit et al., 2011). Each regulator has its own binding requirements and recognizes specific DNA sequences. AfsR is able to bind to the promoters of other Pho regulon members, such as pstS and phoRP (i.e., establishing a nice cross-talk between these two proteins). This kind of interaction may allow streptomycetes to fine-tune secondary metabolism in relation to different signals, like for example Pi scarcity and SAM accumulation. In general, it can be conclude that the effect of the Pi concentration on antibiotic production is much more conserved and severe than the Pho-dependent effect, which means that unknown additional systems may account for the Pi antibiotic production control.
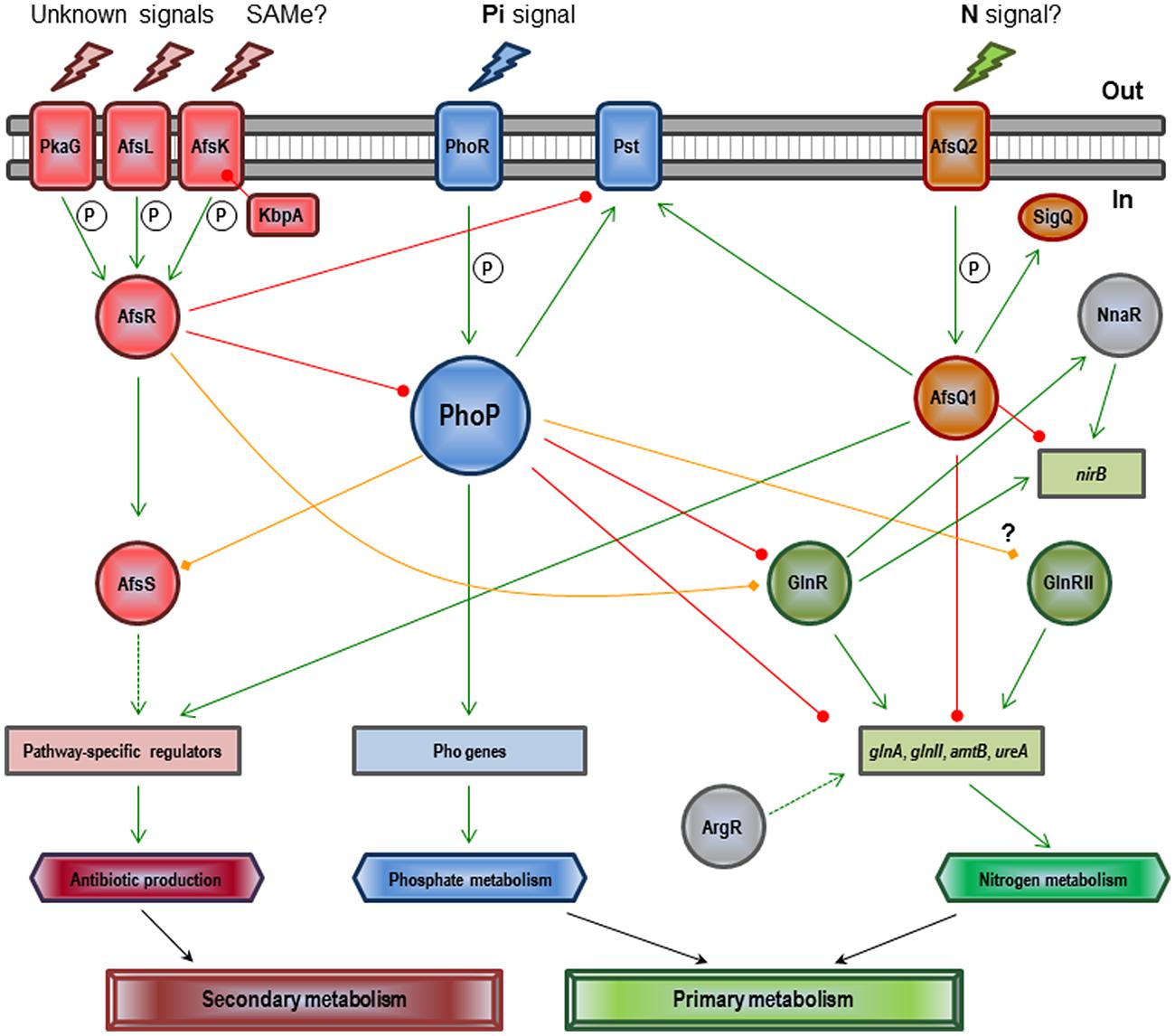
FIGURE 2. Regulatory network involving PhoP and other Streptomyces coelicolor global regulators in the control of Pi and N metabolisms and antibiotic production (see text). Green arrows indicate induction. Circle-ended red lines indicate repression. Rhombus-ended orange lines indicate dual activation/repression or unknown regulation. Dashed lines indicate indirect or uncharacterized links. P indicates activation by phosphorylation. SAMe (S-adenosyl-L-methionine), Pi (inorganic phosphate), and N (nitrogen).
Nutritional Stress Networking
Bacterial systems involved in carbon (C), nitrogen (N), and Pi metabolisms must interact for coordination, but the mechanisms involved in these interactions are just starting to be understood. This section focuses mainly on the interactions of Pi regulation with those of C and N, although other nutritional interconnections are also described (see Table 1).
Pi and C Cross-Talk
Several works have reported the regulation of Pho regulon members by different C sources as well as by different known C regulators, but only some describe this regulation at the molecular level. One of the most interesting studies describes the direct involvement of CCR (carboepression) in the expression of the phoPR operon in B. subtilis (Puri-Taneja et al., 2006). This operon is repressed by direct binding of CcpA (a transcriptional regulator mediating CCR) to a cre box located immediately upstream of the PHO box. In the same direction, Choi and Saier (2005) showed that CcpA influences expression of other Pho regulon genes in B. subtilis such as the alkaline phosphatase genes, phoA, and phoB. These authors showed that the effect of glucose on these two genes depends not only on CcpA but also on the growth conditions, including Pi and N sources. Similar interactions have been also shown in other bacteria, for example Vibrio vulnificus (Oh et al., 2007). Another example of Pi and C interaction is the regulation of the ugp operon in E. coli, which is induced under both Pi- and C-limiting conditions from two separate promoters. The downstream promoter has multiple PHO boxes while the upstream promoter has a consensus sequence for the cAMP–CRP complex, which is formed under the presence of non-preferred carbon sources (Kasahara et al., 1991). A similar control involving these two regulatory pathways has also been described for psiE in E. coli (Kim et al., 2000). In C. glutamicum, the global cAMP-dependent transcriptional regulator, known as GlxR, has been shown to bind to the pstS promoter and activate its transcription under Pi-limiting conditions in a C source-dependent manner (Panhorst et al., 2011). The existence of additional Pho-dependent promoters controlled by the C source has been reported in three S. coelicolor genes: pstS, glpQ1, and glpQ2 (Díaz et al., 2005; Esteban et al., 2008; Santos-Beneit et al., 2009b).
Cross-regulation in the other direction (i.e., Pi to C) has been also documented. For example, a connection between the glucose-Pi stress response (mediated by the SgrR and SgrS regulators) and the Pi metabolism has been reported in E. coli. Direct induction of the Pho regulon is sufficient to partially suppress the glucose-Pi stress growth defect of a sgrS mutant; although the molecular mechanism is yet unclear (Richards and Vanderpool, 2012). Another example of Pi affecting C metabolism has been shown in S. coelicolor in which PhoP is able to repress glycogen catabolism and gluconeogenesis by means of the regulation of key enzymes of these pathways (Thomas et al., 2012). A more complex network involving C and Pi regulations has been described in a recent work with the soil bacterium Rhizobium leguminosarum. In this study, among the different nutrients tested, iron, C, and Pi have been found to strongly affect transcription of the pssA gene (involved in the first step of exopolysaccharide synthesis). The regulation of this gene has been shown to be very complex, involving at least three different promoters and the binding of at least four different regulators, RosR, RirA, cAMP–CRP, and PhoB; responding to flavonoids (and other environmental factors), iron, C, and Pi, respectively, (Janczarek and Urbanik-Sypniewska, 2013). Overall, these results reflect a coordinated redevelopment of metabolism in many different bacteria to balance the availability of Pi with C utilization.
Pi and N Cross-Talk
It has long been known that N sources and Pi content regulate microbial growth through interconnected networks (Doull and Vining, 1990). One of the first interactions described at the molecular level was the link between the glutamate and Pi positive effects over the ykoL gene in B. subtilis. The gene is induced by both PhoP and TnrA regulators (Robichon et al., 2000). TnrA is involved, together with GlnR, CodY, and SigL, in the control of N metabolism in response to N supply (Fisher, 1999). A link between Pi and N regulations has been also described in E. coli. In this bacterium, the transcription of phoB, and many other Pho regulon members is up-regulated under N limitation (Marzan and Shimizu, 2011). On the other hand, putative PHO boxes have been detected in several genes involved in E. coli N metabolism, such as those of the glutamine permease operon glnHPQ and the glnLG two-component system that regulates the glutamine synthetase glnA gene (Merrick and Edwards, 1995; Rodríguez-García et al., 2009).
The interplay between Pi and N regulatory pathways reaches its maximum complexity in S. coelicolor (see Figure 2). Under Pi limitation, PhoP represses transcription of the major N regulator, glnR, as well as that of glnA, glnII, amtB, and ureA (all GlnR targets) by direct binding to their promoters; actually, PhoP and GlnR compete for binding at least in glnA, glnII, and amtB promoters (Tiffert et al., 2008; Rodríguez-García et al., 2009; Wang et al., 2012; Sola-Landa et al., 2013). Interestingly, the PhoP binding site in glnA overlaps not only with GlnR but also with another regulator, AfsQ1; existing three global regulators competing for the same promoter (Wang et al., 2013). AfsQ1 is the response regulator of a two component system (AfsQ2–AfsQ1) which controls antibiotic production and N, C, and Pi uptake (Shu et al., 2009). The network is even more complex: besides GlnR, AfsQ1, and PhoP, the regulators GlnRII, NnaR, AfsR, and ArgR are also involved. GlnRII is a second N regulator which binds to most of the GlnR targets, although there is no agreement as to whether it is controlled by PhoP or not (Rodríguez-García et al., 2009; Allenby et al., 2012). NnaR, in cooperation with GlnR, regulates several N assimilation genes including nirB, which is also controlled by AfsQ1 (Amin et al., 2012; Wang et al., 2013). The secondary metabolite regulator AfsR binds to the promoter region of glnR and overlaps with the binding site of PhoP (Rodríguez-García et al., 2009; Santos-Beneit et al., 2012). Finally, the arginine regulator ArgR is needed for optimal glnA expression (Pérez-Redondo et al., 2012). In general, under conditions of Pi limitation, PhoP represses genes required for N assimilation because it is unnecessary without a supply of cellular Pi.
Pi Cross-Talk with other Nutrient Stresses
Inorganic phosphate cross-talk with other nutrients has been also reported. For example, under low Pi conditions, some cyanobacteria substitute phosphorus with sulfur within their membrane lipids (Van Mooy et al., 2006). In fact, Pi limitation has been shown to increase the transcription of sulfate transporters genes in the cyanobacterium Microcystis aeruginosa (Harke and Gobler, 2013). Another example is the tight co-regulation existing between arsenic and Pi metabolisms in Agrobacterium tumefaciens. This interaction involves the Pi suppression of arsenite oxidation and the arsenite enhancing of Pi stress response (Kang et al., 2012). The physiological explanation for this cross-talk is complex and might involve other mechanisms that allow the microbial cell to react to Pi-limiting situations and avoid the toxic biochemical effects of arsenic. In S. meliloti, several genes coding for iron transporters have been described as members of the Pho regulon (Yuan et al., 2006). Alternatively, iron has been proved to be an important modulator of the activity of Pho-regulated phosphatases in Pseudomonas fluorescens, although this control is not at the transcriptional level (Monds et al., 2006). More interesting is the cross-talk between Pho and Fur systems in the bacterial pathogen Edwardsiella tarda (Chakraborty et al., 2011). In this bacterium, the ferric uptake regulator Fur senses changes in iron concentration and regulates the expression of type III and VI secretion systems through the EsrC regulator. Under low Pi concentrations, PhoB suppresses the secretion, and expression of proteins from the type III and VI secretion systems, either directly or indirectly through esrC control. On the other hand, under high iron concentration, Fur suppresses the transcription of phoB, pstSCAB–phoU, and esrC in an indirect manner via unknown regulators, perhaps involving an interaction between Fur and PhoU proteins (Chakraborty et al., 2011). This cross-talk is of biological significance because iron regulation is crucial for controlling the pathogenicity of many bacteria. Another example of links between nutrients is the influence of sodium in the expression of the phoPR operon of B. subtilis by means of the NhaC regulator. In this bacterium, several genes of the Pho regulon have been shown to be overexpressed in the absence of NhaC (dependent on the external sodium concentration) and repressed when the protein is overproduced, in accordance with the negative effect on growth of NhaC (Prágai et al., 2001). Finally, an interesting link involving at least three different nutrients (potassium, N, and Pi) has been described in E. coli (Lüttmann et al., 2012). In this bacterium, the co-sensing protein EIIANtr, a member of the so-called nitrogen-related phosphotransferase system (PTSNtr), has been shown to modulate the activities of two distinct sensor kinases, KdpD (involved in regulation of potassium uptake) and PhoR. EIIANtr interacts with both proteins and stimulates their kinase activity, as shown by in vivo and in vitro experiments (Lüttmann et al., 2012). The two component system KdpD–KdpE positively controls the synthesis of the high-affinity potassium transporter KdpFABC under low concentration of this nutrient (Lüttmann et al., 2009); and PhoR–PhoP activates the synthesis of the high-affinity Pi transporter PstSCAB under low Pi concentration. Therefore, EIIANtr globally stimulates both high-affinity potassium and Pi transports; although regulations of Kdp and Pho regulons are independent (i.e., potassium and Pi regulations do not cross-talk).
Pi Starvation and General Stresses
Interaction of the Pho regulon with other stresses such as the oxidative, osmotic, acid, and cell wall stresses has been also documented (see next section). For example, several studies in distinct bacteria have reported the Pho-dependent up-regulation of genes coding for catalases that protect bacteria from the oxidative stress by degrading hydrogen peroxide (Yuan et al., 2005). Alternatively, different works have brought to light links between the Pho regulon and the synthesis of polyphosphate and guanosine tetraphosphate (ppGpp). Polyphosphate is a known stress response molecule which is very important for the stationary-phase survival of bacteria and is synthesized in a Pho-dependent manner when environmental stresses occur (Kornberg et al., 1999; Shiba et al., 2000). On the other hand, ppGpp is a key phosphate containing molecule involved in the stringent response, which is responsible for the inhibition of RNA synthesis when there is a deficiency in the amino acids availability (Chatterji and Ojha, 2001). The Pho regulon is repressed in E. coli mutants that cannot accumulate ppGpp, such as relA and spoT mutants, whereas ppGpp content is altered in pst mutants (Loewen et al., 1998; Spira and Yagil, 1998). In addition, some pstA–pstB mutants have been shown to alter the accumulation of RpoS; a sigma factor implicated in the cellular response to many environmental stresses (Schurdell et al., 2007). RpoS is highly unstable in exponentially growing cells, whereas its stability increases dramatically upon ppGpp accumulation (Gentry et al., 1993) and under certain stress conditions such as Pi starvation (Bougdour et al., 2006) or magnesium starvation (Bougdour et al., 2008). The small protein IraP accounts for RpoS stabilization during Pi starvation conditions by blocking the action of RssB, an adaptor protein for RpoS degradation (Bougdour et al., 2006). Interestingly, the iraP promoter is positively regulated by ppGpp (Bougdour and Gottesman, 2007). All these results demonstrate that the Pi cross-talk with nutrient and general stresses is indicative that all stresses regulation might have common features associated to stress responses, for example associated to RpoS regulon, as well as features specific to the applied stress.
The Pho Regulon is Involved in Pathogenesis
Bacterial pathogens, including those of humans, animals, and plants, deal with Pi-limiting or Pi-rich environments in the host depending on the site of infection. Although the main function of the Pho regulon is the control of the Pi homeostasis, this system also plays a role in pathogenesis. However, the mechanisms connecting the Pho regulon with pathogenesis are not always well defined. This section focuses in a small part of the numerous reports that showed a relationship between Pi regulation and pathogenesis. Most of them were performed in V. cholerae, Pseudomonas spp. and pathogenic E. coli species (see reviews of Lamarche et al., 2008; Crépin et al., 2011; Chekabab et al., 2014). There are several strategies accounting for virulence in which the Pho regulon has been shown to be involved. The main ones are: (a) tolerance to acidity, (b) toxin production, (c) biofilm formation, and (d) resistance to antimicrobial compounds.
(a) In some bacteria, the Pho system is able to respond to external acidity by controlling the transcription of genes important for acid shock resistance. One example is the PhoB-dependent activation of the acid-inducible asr gene in E. coli. The Asr protein has a role in protecting periplasmic proteins from the deleterious effects of low pH environments, such as those of the stomach or intestine, indicating a direct role of PhoB in resistance to acidity (Suziedeliene et al., 1999; Gajiwala and Burley, 2000).
(b) One of the clearest examples of the involvement of the Pho regulon on toxin production is reported in V. cholera. In this bacterium, the regulatory cascade leading to the production of TCP and CT toxins is repressed by PhoB under Pi-limiting conditions. However, under the normal high Pi conditions of the gut, the regulatory cascade is not repressed, being able to activate TCP and CT productions (Pratt et al., 2010). Another example has been described in Pseudomonas aeruginosa, although in the opposite direction. In this pathogen, the production of the pyocyanin toxin is positively activated by PhoB. This is not strange, because infection with Gram-negative pathogens normally leads to Pi reduction in the plasma to a suboptimal level for bacterial growth. In this manner, it has been shown that a host supplemented with excess Pi is less susceptible to P. aeruginosa infection than the non-supplemented (Jensen et al., 2006; Zaborin et al., 2012). This result suggests that the opportunistic pathogen P. aeruginosa can use Pi deficiency as an environmental signal of host trauma to shift from non-virulent to lethal phenotypes. In a similar way, it has been shown that activation of the Pho regulon induces a shift of Bacteroides fragilis from gut symbiosis to pathogenicity (Wakimoto et al., 2013).
(c) Biofilms are mixed microbial populations typically embedded in a matrix of extracellular polymers that lead to a number of functions, including: concentration of virulence factors, tolerance to environmental stresses, resistance to antimicrobial agents, redox regulation, nutrition, adhesiveness, locomotion, etc. (Flemming and Wingender, 2010). A link between Pi homeostasis and biofilm formation has been reported in different bacteria. For example, in the plant pathogen A. tumefaciens biofilm formation is enhanced in low Pi conditions by PhoB (Danhorn et al., 2004). Influence of the Pho regulon on biofilm formation has been also described in Pseudomonas, Proteus mirabilis, E. coli, and V. cholerae (Monds et al., 2001, 2007; O’May et al., 2009). In P. aeruginosa, Pi limitation promotes bacterial hyper-swarming through PhoB induction of rhamnolipid biosurfactants (Blus-Kadosh et al., 2013). On the other hand, the published data on enteric pathogens suggest a regulatory model in which biofilm formation would be promoted during the course of infection, whereas under Pi-limiting conditions (such as those of the aquatic environments outside of the host), PhoB would shut down biofilm formation, and promote bacterial survival and dissemination (Pratt et al., 2009; Yoshida et al., 2010).
(d) The lower susceptibility of biofilm-grown bacteria to antimicrobial agents, such as antibiotics and biocides, has been widely investigated (Mah, 2012). However, involvement of Pi regulation in antibiotic resistance by distinct mechanisms to those related with biofilms has been also reported. For example, Pi concentration has been shown to regulate to a high extent vancomycin resistance in two different streptomycetes: S. coelicolor and S. lividans (Santos-Beneit and Martín, 2013). Interestingly, an S. coelicolor mutant in a PhoP down-regulated gene (putatively involved in exopolysaccharide synthesis) has been shown to be at least ten times more resistant to vancomycin than the wild type strain (Santos-Beneit et al., 2014). In Enterococcus faecium, PhoB can be activated by the sensor kinase VanS (the partner of the VanR response regulator, which is involved in the induction of vancomycin resistance genes), pointing to a cross-talk between the Pi starvation response and the vancomycin resistance pathways in this bacterium (Fisher et al., 1995). In other work, it has been shown that pst mutants of pathogenic E. coli strains display an increased sensitivity to vancomycin and different cationic antimicrobial peptides (Lamarche et al., 2008). Something similar has been described in M. tuberculosis when the phoU homolog gene phoY2 is inactivated (Shi and Zhang, 2010).
Therefore, understanding Pi regulation in pathogens and/or in hosts may be of great value for the development of new drugs.
Concluding Remarks (Perspectives)
The physiological response to Pi-starvation in bacteria is a good example for systems biology studies, not only because it constitutes a fast and very sensible responding system, but also because it generates an amazing regulatory network with few precedents in bacteria, involving primary and secondary metabolisms, virulent factors, and multiple cross-talks in some cases. Different transcriptomic analyses and other works have brought to light the following general observations for the Pi regulatory network in bacteria:
(a) In most bacteria, the Pho response controls many more genes than just those related to Pi metabolism; revealing a surprisingly diverse role for the system, not only as a regulator of Pi assimilation, but also as a “master” regulator of the cell.
(b) Many different levels of Pho-dependent regulation are possible, including up-regulation, down-regulation, and complex regulatory patterns. Actually, the number of genes negatively regulated (directly or indirectly) is normally larger than the number of activated genes (Martín et al., 2012). Two different requirements of the response regulator may account for this difference: (i) the response regulator needs to be phosphorylated in order to binding tightly to the DNA and/or form strong dimers and multimers (Allen et al., 2001; Bachhawat et al., 2005). (ii) The response regulator must be able to interact with RNA polymerase in order to activate transcription (Makino et al., 1993; Chen et al., 2004). These requirements are not essential for transcriptional repression, for which a very weak contact can hamper the biding or movement of RNA polymerase or the binding of a transcriptional activator (Browning and Busby, 2004). This difference explains why many genes can be slightly regulated by the Pho system before Pi depletion; although transcriptional activation only occurs under Pi starvation (i.e., when the response regulator is in its phosphorylated state). This fact is clearly observed in V. cholerae, in which the Pho system is required for full growth and normal stress response, not only under Pi limiting conditions but also under Pi abundance (Lery et al., 2013).
(c) The total number of genes involved in the Pi-limitation response is expected to be considerably larger than those regulated just by PhoP, pointing to alternative systems of Pi regulation in bacteria. For example, two Pho-independent systems (one being the σB general stress regulon and the other remaining unknown) have been shown to be involved in the B. subtilis Pi starvation response (Antelmann et al., 2000; Prágai and Harwood, 2002). In addition, up to three two-component systems (PhoR1–PhoP1, PhoR2–PhoP2, and PhoR3–PhoP3) and an orphan response regulator (PhoP4) have been shown to be responsible for the Pi regulation response in the bacterium Myxococcus xanthus (Moraleda-Muñoz et al., 2003; Pham et al., 2006). Finally, two hierarchic response systems to Pi scarcity have been proposed in the marine cyanobacterium Synechococcus sp. WH8102, the first being the typical PhoR–PhoB system and the second depending on the CRP family transcriptional regulator PtrA (Ostrowski et al., 2010).
Advances in the field of Pi regulation promise to be very important for new developments in biotechnology. Therefore, future work should focus on the complete characterization of the mechanism systems involved in bacterial Pi regulation (either novel or depending on the Pho system); investing greater effort on those related to secondary metabolite production control and pathogenesis. These systems may include other players distinct to proteins (or additional), such as small non-coding RNAs, metabolites, and precursors.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by a Wellcome Trust Senior Investigator Award (WT098374AIA) to Professor Jeff Errington. I thank María Ordóñez Robles for making constructive comments about the manuscript. I also thank the Spanish Government for the financial support of all my work in the Streptomyces Pho regulon and colleges who were involved in the project at the Institute of Biotechnology of León (Spain), especially Professor Juan F. Martín and Dr. Antonio Rodríguez-García.
References
Aharonowitz, Y., and Demain, A. L. (1977). Influence of inorganic phosphate and organic buffers on cephalosporin production by Streptomyces clavuligerus. Arch. Microbiol. 115, 169–173. doi: 10.1007/BF00406371
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Allen, M. P., Zumbrennen, K. B., and McCleary, W. R. (2001). Genetic evidence that the alpha5 helix of the receiver domain of PhoB is involved in interdomain interactions. J. Bacteriol. 183, 2204–2211. doi: 10.1128/JB.183.7.2204-2211.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Allenby, N. E., Laing, E., Bucca, G., Kierzek, A. M., and Smith, C. P. (2012). Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic Acids Res. 40, 9543–9556. doi: 10.1093/nar/gks766
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Amemura, M., Makino, K., Shinagawa, H., and Nakata, A. (1990). Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM-open reading frame 2. J. Bacteriol. 172, 6300–6307.
Amin, R., Reuther, J., Bera, A., Wohlleben, W., and Mast, Y. (2012). A novel GlnR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor. Microbiology 158, 1172–1182. doi: 10.1099/mic.0.054817-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Antelmann, H., Harf, C., and Hecker, M. (2000). Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182, 4478–4490. doi: 10.1128/JB.182.16.4478-4490.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Asturias, J. A., Martín, J. F., and Liras, P. (1994). Biosynthesis and phosphate control of candicidin by Streptomyces acrimycini JI2236: effect of amplification of the pabAB gene. J. Ind. Microbiol. 13, 183–189. doi: 10.1007/BF01584005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Auesukaree, C., Homma, T., Tochio, H., Shirakawa, M., Kaneko, Y., and Harashima, S. (2004). Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 279, 17289–17294. doi: 10.1074/jbc.M312202200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bachhawat, P., Swapna, G. V., Montelione, G. T., and Stock, A. M. (2005). Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 13, 1353–1363. doi: 10.1016/j.str.2005.06.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bergwitz, C., and Jüppner, H. (2011). Phosphate sensing. Adv. Chronic. Kidney Dis. 18, 132–144. doi: 10.1053/j.ackd.2011.01.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Birkey, S. M., Liu, W., Zhang, X., Duggan, M. F., and Hulett, F. M. (1998). Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30, 943–953. doi: 10.1046/j.1365-2958.1998.01122.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blanco, A. G., Canals, A., Bernués, J., Solà, M., and Coll, M. (2011). The structure of a transcription activation subcomplex reveals how σ(70) is recruited to PhoB promoters. EMBO J. 30, 3776–3785. doi: 10.1038/emboj.2011.271
Blanco, A. G., Canals, A., and Coll, M. (2012). PhoB transcriptional activator binds hierarchically to pho box promoters. Biol. Chem. 393, 1165–1171. doi: 10.1515/hsz-2012-0230
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blanco, A. G., Solà, M., Gomis-Rüth, F. X., and Coll, M. (2002). Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10, 701–713. doi: 10.1016/S0969-2126(02)00761-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blus-Kadosh, I., Zilka, A., Yerushalmi, G., and Banin, E. (2013). The effect of pstS and phoB on quorum sensing and swarming motility in Pseudomonas aeruginosa. PLoS ONE 8:e74444. doi: 10.1371/journal.pone.0074444
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Botella, E., Devine, S. K., Hubner, S., Salzberg, L. I., Gale, R. T., Brown, E. D., et al. (2014). PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol. Microbiol. 94, 1242–1259. doi: 10.1111/mmi.12833
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bougdour, A., Cunning, C., Baptiste, P. J., Elliott, T., and Gottesman, S. (2008). Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 68, 298–313. doi: 10.1111/j.1365-2958.2008.06146.x
Bougdour, A., and Gottesman, S. (2007). ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc. Natl. Acad. Sci. U.S.A. 104, 12896–12901. doi: 10.1073/pnas.0705561104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bougdour, A., Wickner, S., and Gottesman, S. (2006). Modulating RssB activity: iraP, a novel regulator of σS stability in Escherichia coli. Genes Dev. 20, 884–897. doi: 10.1101/gad.1400306
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Browning, D. F., and Busby, S. J. (2004). The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2, 57–65. doi: 10.1038/nrmicro787
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carmany, D. O., Hollingsworth, K., and McCleary, W. R. (2003). Genetic and biochemical studies of phosphatase activity of PhoR. J. Bacteriol. 185, 1112–1115. doi: 10.1128/JB.185.3.1112-1115.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chakraborty, S., Sivaraman, J., Leung, K. Y., and Mok, Y. K. (2011). Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J. Biol. Chem. 286, 39417–39430. doi: 10.1074/jbc.M111.295188
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chatterji, D., and Ojha, A. K. (2001). Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4, 160–165. doi: 10.1016/S1369-5274(00)00182-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chekabab, S. M., Harel, J., and Dozois, C. M. (2014). Interplay between genetic regulation of phosphate homeostasis and bacterial virulence. Virulence 5, 786–793. doi: 10.4161/viru.29307
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, Y., Abdel-Fattah, W. R., and Hulett, F. M. (2004). Residues required for Bacillus subtilis PhoP DNA binding or RNA polymerase interaction: alanine scanning of PhoP effector domain transactivation loop and alpha helix 3. J. Bacteriol. 186, 1493–1502. doi: 10.1128/JB.186.5.1493-1502.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, Y., Birck, C., Samama, J. P., and Hulett, F. M. (2003). Residue R113 is essential for PhoP dimerization and function: a residue buried in the asymmetric PhoP dimer interface determined in the PhoPN three-dimensional crystal structure. J. Bacteriol. 185, 262–273. doi: 10.1128/JB.185.1.262-273.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cheng, Y. R., Hauck, L., and Demain, A. L. (1995). Phosphate, ammonium, magnesium and iron nutrition of Streptomyces hygroscopicus with respect to rapamycin biosynthesis. J. Ind. Microbiol. 14, 424–427. doi: 10.1007/BF01569962
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Choi, S.-K., and Saier, M. H. (2005). Regulation of pho regulon gene expression by the carbon control protein A, CcpA, in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 10, 40–50. doi: 10.1159/000090347
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chouayekh, H., and Virolle, M. J. (2002). The polyphosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans. Mol. Microbiol. 43, 919–930. doi: 10.1046/j.1365-2958.2002.02557.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Crépin, S., Chekabab, S. M., Le Bihan, G., Bertrand, N., Dozois, C. M., and Harel, J. (2011). The Pho regulon and the pathogenesis of Escherichia coli. Vet. Microbiol. 153, 82–88. doi: 10.1016/j.vetmic.2011.05.043
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danhorn, T., Hentzer, M., Givskov, M., Parsek, M. R., and Fuqua, C. (2004). Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186, 4492–4501. doi: 10.1128/JB.186.14.4492-4501.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Díaz, M., Esteban, A., Fernández-Abalos, J. M., and Santamaría, R. I. (2005). The high-affinity phosphate-binding protein PstS is accumulated under high fructose concentrations and mutation of the corresponding gene affects differentiation in Streptomyces lividans. Microbiology 151, 2583–2592. doi: 10.1099/mic.0.27983-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Doull, J. L., and Vining, L. C. (1990). Nutritional control of actinorhodin production by Streptomyces coelicolor A3(2): suppressive effects of nitrogen and phosphate. Appl. Microbiol. Biotechnol. 32, 449–454. doi: 10.1007/BF00903781
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellison, D. W., and McCleary, W. R. (2000). The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J. Bacteriol. 182, 6592–6597. doi: 10.1128/JB.182.23.6592-6597.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Esteban, A., Díaz, M., Yepes, A., and Santamaría, R. I. (2008). Expression of the pstS gene of Streptomyces lividans is regulated by the carbon source and is partially independent of the PhoP regulator. BMC Microbiol. 8:201. doi: 10.1186/1471-2180-8-201
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fernández-Martínez, L. T., Santos-Beneit, F., and Martín, J. F. (2012). Is PhoR-PhoP partner fidelity strict? PhoR is required for the activation of the pho regulon in Streptomyces coelicolor. Mol. Genet. Genomics 287, 565–573. doi: 10.1007/s00438-012-0698-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fisher, S. H. (1999). Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32, 223–232. doi: 10.1046/j.1365-2958.1999.01333.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fisher, S. L., Jiang, W., Wanner, B. L., and Walsh, C. T. (1995). Crosstalk between the histidine protein kinase VanS and the response regulator PhoB. Characterization and identification of a VanS domain that inhibits activation of PhoB. J. Biol. Chem. 270, 23143–23149. doi: 10.1074/jbc.270.39.23143
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gajiwala, K. S., and Burley, S. K. (2000). HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295, 605–612. doi: 10.1006/jmbi.1999.3347
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gardner, S. G., Johns, K. D., Tanner, R., and McCleary, W. R. (2014). The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J. Bacteriol. 196, 1741–1752. doi: 10.1128/JB.00029-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gentry, D. R., Hernandez, V. J., Nguyen, L. H., Jensen, D. B., and Cashel, M. (1993). Synthesis of the stationary-phase sigma factor S is positively regulated by ppGpp. J. Bacteriol. 175, 7982–7989.
Ghorbel, S., Smirnov, A., Chouayekh, H., Sperandio, B., Esnault, C., Kormanec, J., et al. (2006a). Regulation of ppk expression and in vivo function of Ppk in Streptomyces lividans TK24. J. Bacteriol. 188, 6269–6276. doi: 10.1128/JB.00202-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ghorbel, S., Kormanec, J., Artus, A., and Virolle, M. J. (2006b). Transcriptional studies and regulatory interactions between the phoR-phoP operon and the phoU, mtpA and ppk genes of Streptomyces lividans TK24. J. Bacteriol. 188, 677–686. doi: 10.1128/JB.188.2.677-686.2006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Glover, R. T., Kriakov, J., Garforth, S. J., Baughn, A. D., and Jacobs, W. R. Jr. (2007). The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189, 5495–5503. doi: 10.1128/JB.00190-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ha, S., and Tran, L. S. (2014). Understanding plant responses to phosphorus starvation for improvement of plant tolerance to phosphorus deficiency by biotechnological approaches. Crit. Rev. Biotechnol. 34, 16–30. doi: 10.3109/07388551.2013.783549
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Harke, M. J., and Gobler, C. J. (2013). Global transcriptional responses of the toxic cyanobacterium, Microcystis aeruginosa, to nitrogen stress, phosphorus stress, and growth on organic matter. PLoS ONE 8:e69834. doi: 10.1371/journal.pone.0069834
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hege-Treskatis, D., King, R., Wolf, H., and Gilles, E. D. (1992). Nutritional control of nikkomycin and juglomycin production by Streptomyces tendae in continuous culture. Appl. Microbiol. Biotechnol. 36, 440–445. doi: 10.1007/BF00170179
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hulett, F. M. (1996). The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19, 933–939. doi: 10.1046/j.1365-2958.1996.421953.x
Hulett, F. M., Lee, J., Shi, L., Sun, G., Chesnut, R., Sharkova, E., et al. (1994). Sequential action of two-component genetic switches regulates the Pho regulon in Bacillus subtilis. J. Bacteriol. 176, 1348–1358.
Jakeman, D. L., Graham, C. L., Young, W., and Vining, L. C. (2006). Culture conditions improving the production of jadomycin B. J. Ind. Microbiol. Biotechnol. 33, 767–772. doi: 10.1007/s10295-006-0113-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Janczarek, M., and Urbanik-Sypniewska, T. (2013). Expression of the Rhizobium leguminosarum bv. trifolii pssA gene, involved in exopolysaccharide synthesis, is regulated by RosR, phosphate, and the carbon source. J. Bacteriol. 195, 3412–3423. doi: 10.1128/JB.02213-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jensen, V., Löns, D., Zaoui, C., Bredenbruch, F., Meissner, A., Dieterich, G., et al. (2006). RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 188, 8601–8606. doi: 10.1128/JB.01378-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jin, Y. Y., Cheng, J., Yang, S. H., Meng, L., Palaniyandi, S. A., Zhao, X. Q., et al. (2011). S-adenosyl-L-methionine activates actinorhodin biosynthesis by increasing autophosphorylation of the Ser/Thr protein kinase AfsK in Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem. 75, 910–913. doi: 10.1271/bbb.100873
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Juntarajumnong, W., Hirani, T. A., Simpson, J. M., Incharoensakdi, A., and Eaton-Rye, J. J. (2007). Phosphate sensing in Synechocystis sp. PCC 6803: SphU and the SphS-SphR two-component regulatory system. Arch. Microbiol. 188, 389–402. doi: 10.1007/s00203-007-0259-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kang, Y. S., Heinemann, J., Bothner, B., Rensing, C., and McDermott, T. R. (2012). Integrated co-regulation of bacterial arsenic and phosphorus metabolisms. Environ. Microbiol. 14, 3097–3109. doi: 10.1111/j.1462-2920.2012.02881.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kasahara, M., Makino, K., Amemura, M., Nakata, A., and Shinagawa, H. (1991). Dual regulation of the ugp operon by phosphate and carbon starvation at two interspaced promoters. J. Bacteriol. 173, 549–558.
Kaushal, B., Paul, S., and Hulett, F. M. (2010). Direct regulation of Bacillus subtilis phoPR transcription by transition state regulator ScoC. J. Bacteriol. 192, 3103–3113. doi: 10.1128/JB.00089-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, S. K., Kimura, S., Shinagawa, H., Nakata, A., Lee, K. S., Wanner, B. L., et al. (2000). Dual transcriptional regulation of the Escherichia coli phosphate-starvation-inducible psiE gene of the phosphate regulon by PhoB and the cyclic AMP (cAMP)-cAMP receptor protein complex. J. Bacteriol. 182, 5596–5599. doi: 10.1128/JB.182.19.5596-5599.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kocan, M., Schaffer, S., Ishige, T., Sorger-Herrmann, U., Wendisch, V. F., and Bott, M. (2006). Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188, 724–732. doi: 10.1128/JB.188.2.724-732.2006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kornberg, A., Rao, N. N., and Ault-Riche, D. (1999). Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68, 89–125. doi: 10.1146/annurev.biochem.68.1.89
Lamarche, M. G., Wanner, B. L., Crépin, S., and Harel, J. (2008). The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473. doi: 10.1111/j.1574-6976.2008.00101.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, P.-C., Umeyama, T., and Horinouchi, S. (2002). afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 43, 1413–1430. doi: 10.1046/j.1365-2958.2002.02840.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lery, L. M., Goulart, C. L., Figueiredo, F. R., Verdoorn, K. S., Einicker-Lamas, M., Gomes, F. M., et al. (2013). A comparative proteomic analysis of Vibrio cholerae O1 wild-type cells versus a phoB mutant showed that the PhoB/PhoR system is required for full growth and rpoS expression under inorganic phosphate abundance. J. Proteomics 86, 1–15. doi: 10.1016/j.jprot.2013.04.038
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, W., Eder, S., and Hulett, F. M. (1998). Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP-P. J. Bacteriol. 180, 753–758.
Loewen, P. C., Hu, B., Strutinsky, J., and Sparling, R. (1998). Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44, 707–717. doi: 10.1139/cjm-44-8-707
Lüttmann, D., Göpel, Y., and Görke, B. (2012). The phosphotransferase protein EIIA (Ntr) modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli. Mol. Microbiol. 86, 96–110. doi: 10.1111/j.1365-2958.2012.08176.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lüttmann, D., Heermann, R., Zimmer, B., Hillmann, A., Rampp, I. S., Jung, K., et al. (2009). Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIANtr in Escherichia coli. Mol. Microbiol. 72, 978–994. doi: 10.1111/j.1365-2958.2009.06704.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mah, T. F. (2012). Biofilm-specific antibiotic resistance. Future Microbiol. 7, 1061–1072. doi: 10.2217/fmb.12.76
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Makino, K., Amemura, M., Kim, S. K., Nakata, A., and Shinagawa, H. (1993). Role of the sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 7, 149–160. doi: 10.1101/gad.7.1.149
Makino, K., Shinagawa, H., Amemura, M., Kimura, S., Nakata, A., and Ishihama, A. (1988). Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 203, 85–95. doi: 10.1016/0022-2836(88)90093-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martin, J. F. (2004). Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR–PhoP system: an unfinished story. J. Bacteriol. 186, 5197–5201. doi: 10.1128/JB.186.16.5197-5201.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martin, J. F., and Demain, A. L. (1976). Control by phosphate of candicidin production. Biochem. Biophys. Res. Commun. 71, 1103–1109. doi: 10.1016/0006-291X(76)90767-1
Martín, J. F., Santos-Beneit, F., Rodríguez-García, A., Sola-Landa, A., Smith, M. C., Ellingsen, T. E., et al. (2012). Transcriptomic studies of phosphate control of primary and secondary metabolism in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 95, 61–75. doi: 10.1007/s00253-012-4129-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martínez-Castro, M., Salehi-Najafabadi, Z., Romero, F., Pérez-Sanchiz, R., Fernández-Chimeno, R. I., Martín, J. F., et al. (2013). Taxonomy and chemically semi-defined media for the analysis of the tacrolimus producer Streptomyces tsukubaensis. Appl. Microbiol. Biotechnol. 97, 2139–2152. doi: 10.1007/s00253-012-4364-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marzan, L. W., and Shimizu, K. (2011). Metabolic regulation of Escherichia coli and its phoB and phoR genes knockout mutants under phosphate and nitrogen limitations as well as at acidic condition. Microb. Cell. Fact. 10, 39. doi: 10.1186/1475-2859-10-39
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Masuma, R., Tanaka, Y., Tanaka, H., and Omura, S. (1986). Production of nanaomycin and other antibiotics by phosphate-depressed fermentation using phosphate-trapping agents. J. Antibiot. (Tokyo). 39, 1557–1564. doi: 10.7164/antibiotics.39.1557
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Matsumoto, A., Hong, S. K., Ishizuka, H., Horinouchi, S., and Beppu, T. (1994). Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene 146, 47–56. doi: 10.1016/0378-1119(94)90832-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McDowall, K. J., Thamchaipenet, A., and Hunter, I. S. (1999). Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding sites of the OmpR family. J. Bacteriol. 181, 3025–3032.
Mendes, M. V., Tunca, S., Antón, N., Recio, E., Sola-Landa, A., Aparicio, J. F., et al. (2007). The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metabol. Eng. 9, 217–227. doi: 10.1016/j.ymben.2006.10.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Merrick, M. J., and Edwards, R. A. (1995). Nitrogen control in bacteria. Microbiol. Rev. 59, 604–622.
Monds, R. D., Newell, P. D., Gross, R. H., and O’Toole, G. A. (2007). Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63, 656–679. doi: 10.1111/j.1365-2958.2006.05539.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Monds, R. D., Newell, P. D., Schwartzman, J. A., and O’Toole, G. A. (2006). Conservation of the Pho regulon in Pseudomonas fluorescens Pf0-1. Appl. Environ. Microbiol. 72, 1910–1924. doi: 10.1128/AEM.72.3.1910-1924.2006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Monds, R. D., Silby, M. W., and Mahanty, H. K. (2001). Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol. Microbiol. 42, 415–426. doi: 10.1046/j.1365-2958.2001.02641.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Moraleda-Muñoz, A., Carrero-Lérida, J., Pérez, J., and Muñoz-Dorado, J. (2003). Role of two novel two-component regulatory systems in development and phosphatase expression in Myxococcus xanthus. J. Bacteriol. 185, 1376–1383. doi: 10.1128/JB.185.4.1376-1383.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Muda, M., Rao, N. N., and Torriani, A. (1992). Role of PhoU in phosphate transport and alkaline phosphatase regulation. J. Bacteriol. 174, 8057–8064.
Müller, P. J., Haubold, G., Menner, M., Grosse, H. H., Ozegowski, J. H., and Bocker, H. (1984). Effect of phosphate on the biosynthesis of nourseothricin by Streptomyces noursei JA 3890b. Z. Allg. Mikrobiol. 24, 555–564. doi: 10.1002/jobm.3630240812
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Novak, R., Cauwels, A., Charpentier, E., and Tuomanen, E. (1999). Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J. Bacteriol. 181, 1126–1133.
Oh, W.-S., Im, Y.-S., Yeon, K.-Y., Yoon, Y.-J., and Kim, J.-W. (2007). Phosphate and carbon source regulation of alkaline phosphatase and phospholipase in Vibrio vulnificus. J. Microbiol. 45, 311–317.
O’May, G. A., Jacobsen, S. M., Longwell, M., Stoodley, P., Mobley, H. L., and Shirtliff, M. E. (2009). The high-affinity phosphate transporter Pst in Proteus mirabilis HI4320 and its importance in biofilm formation. Microbiology 155, 1523–1535. doi: 10.1099/mic.0.026500-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ostrowski, M., Mazard, S., Tetu, S. G., Phillippy, K., Johnson, A., Palenik, B., et al. (2010). PtrA is required for coordinate regulation of gene expression during phosphate stress in a marine Synechococcus. ISME J. 4, 908–921. doi: 10.1038/ismej.2010.24
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Panhorst, M., Sorger-Herrmann, U., and Wendisch, V. F. (2011). The pstSCAB operon for phosphate uptake is regulated by the global regulator GlxR in Corynebacterium glutamicum. J. Biotechnol. 154, 149–155. doi: 10.1016/j.jbiotec.2010.07.015
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Paul, S., Birkey, S., Liu, W., and Hulett, F. M. (2004). Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP. J. Bacteriol. 186, 4262–4275. doi: 10.1128/JB.186.13.4262-4275.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pérez-Redondo, R., Rodríguez-García, A., Botas, A., Santamarta, I., Martín, J. F., and Liras, P. (2012). ArgR of Streptomyces coelicolor is a versatile regulator. PLoS ONE 7:e32697. doi: 10.1371/journal.pone.0032697
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pham, V. D., Shebelut, C. W., Jose, I. R., Hodgson, D. A., Whitworth, D. E., and Singer, M. (2006). The response regulator PhoP4 is required for late developmental events in Myxococcus xanthus. Microbiology 152, 1609–1620. doi: 10.1099/mic.0.28820-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Prágai, Z., Allenby, N. E., O’Connor, N., Dubrac, S., Rapoport, G., Msadek, T., et al. (2004). Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 186, 1182–1190. doi: 10.1128/JB.186.4.1182-1190.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Prágai, Z., Eschevins, C., Bron, S., and Harwood, C. R. (2001). Bacillus subtilis NhaC, an Na+/H+ antiporter, influences expression of the phoPR operon and production of alkaline phosphatases. J. Bacteriol. 183, 2505–2515. doi: 10.1128/JB.183.8.2505-2515.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Prágai, Z., and Harwood, C. R. (2002). Regulatory interactions between the Pho and sigma (B)-dependent general stress regulons of Bacillus subtilis. Microbiology 148, 1593–1602.
Pratt, J. T., Ismail, A. M., and Camilli, A. (2010). PhoB regulates both environmental and virulence gene expression in Vibrio cholerae. Mol. Microbiol. 77, 1595–1605. doi: 10.1111/j.1365-2958.2010.07310.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pratt, J. T., McDonough, E., and Camilli, A. (2009). PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J. Bacteriol. 191, 6632–6642. doi: 10.1128/JB.00708-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Puri-Taneja, A., Paul, S., Chen, Y., and Hulett, F. M. (2006). CcpA causes repression of the phoPR promoter through a novel transcription start site, P (A6). J. Bacteriol. 188, 1266–1278. doi: 10.1128/JB.188.4.1266-1278.2006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rice, C. D., Pollard, J. E., Lewis, Z. T., and McCleary, W. R. (2009). Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl. Environ. Microbiol. 75, 573–582. doi: 10.1128/AEM.01046-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Richards, G. R., and Vanderpool, C. K. (2012). Induction of the Pho regulon suppresses the growth defect of an Escherichia coli sgrS mutant, connecting phosphate metabolism to the glucose-phosphate stress response. J. Bacteriol. 194, 2520–2530. doi: 10.1128/JB.00009-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Robichon, D., Arnaud, M., Gardan, R., Pragai, Z., O’Reilly, M., Rapoport, G., et al. (2000). Expression of a new operon from Bacillus subtilis, ykzB-ykoL, under the control of the TnrA and PhoP-PhoR global regulators. J. Bacteriol. 182, 1226–1231. doi: 10.1128/JB.182.5.1226-1231.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rodríguez-García, A., Sola-Landa, A., Apel, K., Santos-Beneit, F., and Martín, J. F. (2009). Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 37, 3230–3242. doi: 10.1093/nar/gkp162
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santos-Beneit, F., Fernández-Martínez, L. T., Rodríguez-García, A., Martín-Martín, S., Ordóñez-Robles, M., Yagüe, P., et al. (2014). Transcriptional response to vancomycin in a highly vancomycin-resistant Streptomyces coelicolor mutant. Future Microbiol. 9, 603–622. doi: 10.2217/fmb.14.21
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santos-Beneit, F., and Martín, J. F. (2013). Vancomycin resistance in Streptomyces coelicolor is phosphate-dependent but is not mediated by the PhoP regulator. J. Global Antimicrob. Resist. 1, 109–113. doi: 10.1016/j.jgar.2013.03.003
Santos-Beneit, F., Rodríguez-García, A., Franco-Domínguez, E., and Martín, J. F. (2008). Phosphate-dependent regulation of the low- and high-affinity transport systems in the model actinomycete Streptomyces coelicolor. Microbiology 154, 2356–2370. doi: 10.1099/mic.0.2008/019539-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santos-Beneit, F., Rodríguez-García, A., and Martín, J. F. (2011). Complex transcriptional control of the antibiotic regulator afsS in Streptomyces: PhoP and AfsR are overlapping, competitive activators. J. Bacteriol. 193, 2242–2251. doi: 10.1128/JB.01462-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santos-Beneit, F., Rodríguez-García, A., and Martín, J. F. (2012). Overlapping binding of PhoP and AfsR to the promoter region of glnR in Streptomyces coelicolor. Microbiol. Res. 167, 532–535. doi: 10.1016/j.micres.2012.02.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santos-Beneit, F., Rodríguez-García, A., Sola-Landa, A., and Martín, J. F. (2009a). Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72, 53–68. doi: 10.1111/j.1365-2958.2009.06624.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santos-Beneit, F., Rodríguez-García, A., Apel, A. K., and Martín, J. F. (2009b). Phosphate and carbon source regulation of two PhoP-dependent glycerophosphodiester phosphodiesterase genes of Streptomyces coelicolor. Microbiology 155, 1800–1811. doi: 10.1099/mic.0.026799-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sawai, R., Suzuki, A., Takano, Y., Lee P.-C., and Horinouchi, S. (2004). Phosphorylation of AfsR by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). Gene 334, 53–61. doi: 10.1016/j.gene.2004.02.046
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schaaf, S., and Bott, M. (2007). Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 189, 5002–5011. doi: 10.1128/JB.00121-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schau, M., Eldakak, A., and Hulett, F. M. (2004). Terminal oxidases are essential to bypass the requirement for ResD for full Pho induction in Bacillus subtilis. J. Bacteriol. 186, 8424–8432. doi: 10.1128/JB.186.24.8424-8432.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schurdell, M. S., Woodbury, G. M., and McCleary, W. R. (2007). Genetic evidence suggests that the intergenic region between pstA and pstB plays a role in the regulation of rpoS translation during phosphate limitation. J. Bacteriol. 189, 1150–1153. doi: 10.1128/JB.01482-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shi, W., and Zhang, Y. (2010). PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 1237–1242. doi: 10.1093/jac/dkq103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shiba, T., Tsutsumi, K., Ishige, K., and Noguchi, T. (2000). Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochemistry 65, 315–323.
Shu, D., Chen, L., Wang, W., Yu Z., Ren, C., Zhang, W., et al. (2009). afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 81, 1149–1160. doi: 10.1007/s00253-008-1738-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Solá, M., Gomis-Rüth, F. X., Serrano, L., González, A., and Coll, M. (1999). Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J. Mol. Biol. 285, 675–687. doi: 10.1006/jmbi.1998.2326
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sola-Landa, A., Moura, R. S., and Martín, J. F. (2003). The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U.S.A. 100, 6133–6138. doi: 10.1073/pnas.0931429100
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sola-Landa, A., Rodríguez-García, A., Amin, R., Wohlleben, W., and Martín, J. F. (2013). Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor. Nucleic Acids Res. 41, 1767–1782. doi: 10.1093/nar/gks1203
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sola-Landa, A., Rodríguez-García, A., Apel, A. K., and Martín, J. F. (2008). Target genes and structure of the direct repeats in the DNA binding sequences of the response regulator PhoP in Streptomyces coelicolor. Nucleic Acids Res. 36, 1358–1368. doi: 10.1093/nar/gkm1150
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spira, B., and Yagil, E. (1998). The relation between ppGpp and the PHO regulon in Escherichia coli. Mol. Gen. Genet. 257, 469–477. doi: 10.1007/s004380050671
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Steed, P. M., and Wanner, B. L. (1993). Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175, 6797–6809.
Strauch, M., Webb, V., Spiegelman, G., and Hoch, J. A. (1990). The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U.S.A. 87, 1801–1805. doi: 10.1073/pnas.87.5.1801
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sun, G., Birkey, S. M., and Hulett, F. M. (1996). Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19, 941–948. doi: 10.1046/j.1365-2958.1996.422952.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Surin, B. P., Dixon, N. E., and Rosenberg, H. (1986). Purification of the phoU protein, a negative regulator of the pho regulon of Escherichia coli K-12. J. Bacteriol. 168, 631–635.
Suziedeliene, E., Suziedelis, K., Garbenciute, V., and Normark, S. (1999). The acid-inducible asr gene in Escherichia coli: transcriptional control by the phoBR operon. J. Bacteriol. 181, 2084–2093.
Tanaka, A., Takano, Y., Ohnishi, Y., and Horinouchi, S. (2007). AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J. Mol. Biol. 369, 322–333. doi: 10.1016/j.jmb.2007.02.096
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tenconi, E., Jourdan, S., Motte, P., Virolle, M. J., and Rigali, S. (2012). Extracellular sugar phosphates are assimilated by Streptomyces in a PhoP-dependent manner. Antonie Van Leeuwenhoek 102, 425–433. doi: 10.1007/s10482-012-9763-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thomas, L., Hodgson, D. A., Wentzel, A., Nieselt, K., Ellingsen, T. E., Moore, J., et al. (2012). Metabolic switches and adaptations deduced from the proteomes of Streptomyces coelicolor wild type and phoP mutant grown in batch culture. Mol. Cell. Proteomics 11:M111.013797. doi: 10.1074/mcp.M111.013797
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tiffert, Y., Supra, P., Wurm, R., Wohlleben, W., Wagner, R., and Reuther, J. (2008). The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67, 861–880. doi: 10.1111/j.1365-2958.2007.06092.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tommassen, J., de Geus, P., Lugtenberg, B., Hackett, J., and Reeves, P. (1982). Regulation of the pho regulon of Escherichia coli K-12. Cloning of the regulatory genes phoB and phoR and identification of their gene products. J. Mol. Biol. 157, 265–274. doi: 10.1016/0022-2836(82)90233-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Umeyama, T., Lee, P. C., and Horinouchi, S. (2002). Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl. Microbiol. Biotechnol. 59, 419–425. doi: 10.1007/s00253-002-1045-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Mooy, B. A. S., Rocap, G., Fredricks, H. F., Evans, C. T., and Devol, A. H. (2006). Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marne environments. Proc. Natl. Acad. Sci. U.S.A. 103, 8607–8612. doi: 10.1073/pnas.0600540103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vu-Trong, K., Bhuwapathanapun, S., and Gray, P. P. (1981). Metabolic regulation in tylosin-producing Streptomyces fradiae: phosphate control of tylosin biosynthesis. Antimicrob. Agents Chemother. 19, 209–212. doi: 10.1128/AAC.19.2.209
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wakimoto, S., Nakayama-Imaohji, H., Ichimura, M., Morita, H., Hirakawa, H., Hayashi, T., et al. (2013). PhoB regulates the survival of Bacteroides fragilis in peritoneal abscesses. PLoS ONE 8:e53829. doi: 10.1371/journal.pone.0053829
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, R., Mast, Y., Wang, J., Zhang, W., Zhao, G., Wohlleben, W., et al. (2013). Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol. Microbiol. 87, 30–48. doi: 10.1111/mmi.12080
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wanner, B. L., and Chang, B. D. (1987). The phoBR operon in Escherichia coli K-12. J. Bacteriol. 169, 5569–5574.
Wösten, M. M., Parker, C. T., van Mourik, A., Guilhabert, M. R., van Dijk, L., and van Putten, J. P. (2006). The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62, 278–291. doi: 10.1111/j.1365-2958.2006.05372.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yoshida, Y., Sugiyama, S., Oyamada, T., Yokoyama, K., and Makino, K. (2010). Identification and characterization of novel phosphate regulon genes, ecs0540-ecs0544, in Escherichia coli O157: H7. Mol. Genet. Genomics 284, 197–205. doi: 10.1007/s00438-010-0559-y
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yoshida, Y., Sugiyama, S., Oyamada, T., Yokoyama, K., and Makino, K. (2012). Novel members of the phosphate in Escherichia coli O157: H7 identified using a whole-genome shotgun approach. Gene 502, 27–35. doi: 10.1016/j.gene.2012.03.064
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuan, Z. C., Zaheer, R., and Finan, T. M. (2005). Phosphate limitation induces catalase expression in Sinorhizobium meliloti, Pseudomonas aeruginosa and Agrobacterium tumefaciens. Mol. Microbiol. 58, 877–894. doi: 10.1111/j.1365-2958.2005.04874.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuan, Z. C., Zaheer, R., Morton, R., and Finan, T. M. (2006). Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 34, 2686–2697. doi: 10.1093/nar/gkl365
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zaborin, A., Gerdes, S., Holbrook, C., Liu, D. C., Zaborina, O. Y., and Alverdy, J. C. (2012). Pseudomonas aeruginosa overrides the virulence inducing effect of opioids when it senses an abundance of phosphate. PLoS ONE 7:e34883. doi: 10.1371/journal.pone.0034883
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: phosphate, Pho regulon, PHO box, regulatory networks, pathogenesis, secondary metabolism
Citation: Santos-Beneit F (2015) The Pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6:402. doi: 10.3389/fmicb.2015.00402
Received: 10 March 2015; Paper pending published: 30 March 2015;
Accepted: 17 April 2015; Published online: 30 April 2015
Edited by:
Inês A. Cardoso Pereira, Instituto de Tecnologia Quimica e Biologica/Universidade Nova de Lisboa, PortugalReviewed by:
Josée Harel, Université de Montréal, CanadaWilliam R. McCleary, Brigham Young University, USA
Copyright © 2015 Santos-Beneit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernando Santos-Beneit, Centre for Bacterial Cell Biology, Institute for Cell and Molecular Biosciences, Baddiley-Clark Building, Medical School, Newcastle University, Richardson Road, Newcastle upon Tyne, NE2 4AX, UK, fernando.santos-beneit@ncl.ac.uk
 Fernando Santos-Beneit
Fernando Santos-Beneit