Corrigendum: Novel Variant of New Delhi Metallo-β-lactamase, NDM-20, in Escherichia coli
- 1Beijing Key Laboratory of Detection Technology for Animal-Derived Food Safety, College of Veterinary Medicine, China Agricultural University, Beijing, China
- 2Key Laboratory of Genetics & Molecular Medicine of Shenzhen, Shenzhen Center for Disease Control and Prevention, Shenzhen, China
The spread of carbapenem-resistant Enterobacteriaceae (CRE) mediated by New Delhi metallo-β-lactamase (NDM) poses a serious challenge to clinicians and has become a major public health concern. NDM has been evolving into variants that possess different hydrolysis activity toward antibiotics, so as to affect treatment strategy. In addition, very few studies on NDM variants have focused on animal-derived bacterial isolates. Our study reports a novel NDM variant, NDM-20, in an isolate of Escherichia coli CCD1 recovered from the food animal swine in China. The isolate that was assigned to ST1114, exhibited high level resistance to all β-lactams tested, including aztreonam and carbapenems. The gene of blaNDM-20 was located on an IncX3-type plasmid, surrounded by multiple insertion sequences. Sequencing analysis demonstrated that blaNDM-20 contained three point mutations at positions 262 (G→T), 460 (A→C), and 809 (G→A), compared with blaNDM-1, and just one point mutation at position 809 (G→A), relative to blaNDM-5. Functional analysis revealed that the blaNDM-20 transformant, DH5α+pHSG398/NDM-20, exhibited a higher resistance to ertapenem than that of blaNDM-1 transformant DH5α+pHSG398/NDM-1. Kinetic parameter analysis showed that NDM-20 had increased enzymatic activity against some penicillins and cephalosporins but decreased carbapenemase activity relative to NDM-5. The identification of NDM-20 further confirms the evolution and prevalence of NDM variants in bacteria of food-animal origin.
Introduction
The global dissemination of carbapenem-resistant Enterobacteriaceae (CRE) has become a major threat to public health. With the increasing global prevalence of New Delhi metallo-β-lactamase (NDM)-producing bacteria, which are highly disseminated in China (Khan et al., 2017), available antibiotics are becoming less effective in the treatment of drug-resistant bacterial infections. Since NDM was first reported in India (Yong et al., 2009), the sequences of 18 NDM variants (NDM-19 unpublished) have been reported. NDMs may be evolving diverse hydrolytic activity toward β-lactams, resulting from amino acid substitutions or insertions (Khan et al., 2017), subsequently, it may affect treatment strategy. In addition, these bacteria producing NDM variants are disseminated not only among humans, but also in food animals (Wang et al., 2017). Even though food-producing animals serve as the major reservoir of NDM-producing bacteria (Madec et al., 2017), apart from our previous description of NDM-17 (Liu et al., 2017), studies on NDM variants of animal-derived bacterial isolates are limited. So, it is important and necessary to monitor NDM evolution in food animals. Here, a novel NDM variant, NDM-20, was identified in an Escherichia coli strain isolated from swine, and was subjected to in-depth molecular characterization.
Materials and Methods
Isolation and Identification of the Bacterial Strain and Phenotypic Screening for MBL Production
To better understand the prevalence of CRE, a routine surveillance of antimicrobial resistance in bacteria of food-producing animal origin was carried out in Shandong province, China, in 2016. The swine anal swab samples were collected and enriched in 5 mL of brain heart infusion broth before being plated onto CHROMagar KPC medium (CHROMagar, Paris, France) for CRE selection. Then, the resistant strains were identified on MHA medium containing 1 mg/L of meropenem. Strain CCD1 was isolated from a swine fecal swab sample from a commercial farm. Identification of the bacterial species was performed by MALDI-TOF MS (Bruker Daltonik, Bremen, Germany) and 16S rRNA sequencing, as described previously (Wang et al., 2013). To detect carbapenemase production, the modified Hodge test was carried out using imipenem and meropenem disks, and for MBL production test, the combined-disk was performed by imipenem with and without EDTA.
Antimicrobial Susceptibility Testing and Detection of Antibiotic Resistance Genes
Antimicrobial susceptibility testing of isolates was carried out using the broth microdilution method, and the results in triplicate with best repeatability were interpreted according to the CLSI and EUCAST (version 7.1) guidelines (Clinical and Laboratory Standards Institute [CLSI], 2015; EUCAST, 2017). The tested isolates and antimicrobial agents are listed in Table 1, and E. coli ATCC 25922 was used as a quality control strain. Carbapenemase-encoding genes blaKPC, blaV IM, blaIMP, blaNDM, blaOXA-23-like, blaOXA-58-like, blaOXA-48-like, blaIND, blaKHM, blaDIM, blaGIM, blaSIM, and blaSPM, and β-lactamase-encoding genes blaTEM, blaSHV, and blaCTX-M were screened as previously described (Dallenne et al., 2010; Poirel et al., 2011; Zhang et al., 2017b), and blaNDM was screened by primer preA and preB in Supplementary Table S1. PCR amplicons were sequenced and then analyzed against the GenBank database.
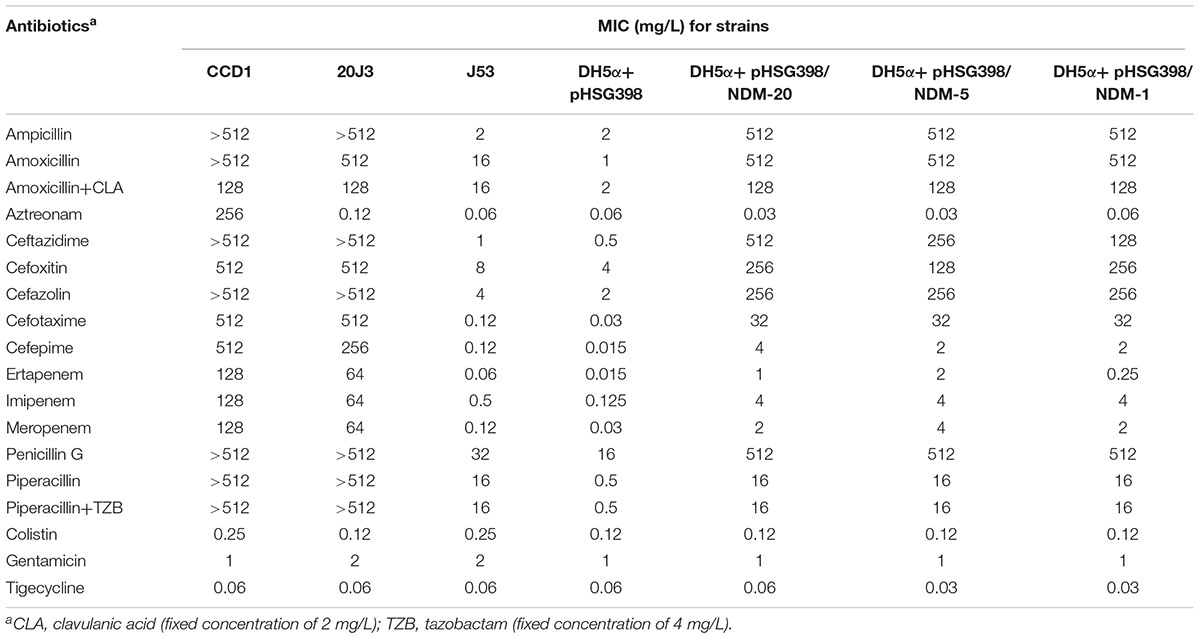
TABLE 1. The resistance gene(s) in CCD1 and β-lactam MICs for the New Delhi metallo-β-lactamase (NDM)-carrying Escherichia coli clinical isolate, transconjugants, and transformants.
Conjugation Assay and Location Analysis of blaNDM
To determine the transferability of blaNDM, transconjugation was attempted using E. coli J53 as the recipient (Liu et al., 2017). Transconjugants were selected on MacConkey agar supplemented with 100 mg/L sodium azide and 1 mg/L meropenem, and confirmed by PCR and sequencing. The transfer frequency was determined from the ratio of transconjugants: donors. Location analysis of blaNDM was performed by S1-PFGE and Southern blotting, as described previously (Borgia et al., 2012).
Resistome and Plasmid Analysis
To further confirm the resistance gene profile of strain CCD1 and to determine the genetic background of blaNDM, the genomic DNA of strain CCD1, including plasmid pNDM-20, was extracted and subjected to WGS, as reported previously (Liu et al., 2017; Wang et al., 2017). A 250-bp paired-end library was constructed using a NEXT Ultra DNA Library Prep kit (New England Biolabs, E7645L) and an Illumina Hiseq 2500 system (Bionova Biotech, Co., Beijing, China). The raw sequencing data were then de novo assembled and examined for the presence of resistance genes using CLC Genomics Workbench 9.0 (CLC Bio, Aarhus, Denmark) against resistance gene database based on CARD. SRST2 was used for MLST analysis of strain CCD1 using the sequence reads (Inouye et al., 2014).
Cloning of blaNDM Genes and Purification of the NDM Proteins
To compare the resistance abilities of blaNDM-20, blaNDM-5 and blaNDM-1, the entire blaNDM-20, blaNDM-5, and blaNDM-1 ORFs were cloned into pHSG398, resulting in recombinant plasmids pHSG398/NDM-20, pHSG398/NDM-5, and pHSG398/NDM-1, respectively (Liu et al., 2017). The plasmids were then transformed into E. coli DH5α. To assay the effect on enzymatic activity caused by Arg270His, blaNDM-20 and blaNDM-5 minus their signal peptide regions were also cloned into the pET28a expression vector, and the resulting plasmids were transformed into E. coli BL21 (DE3) (TransGen Biotech, Beijing, China) (Liu et al., 2017). The corresponding primers were described in Supplementary Table S1. All the transformants were confirmed by PCR and sequencing analysis, and their sensitivity to β-lactams were determined as described above. Purification and identification of the NDM proteins was carried out as per the manufacturer’s instructions in the following order: (1) purification of recombinant NDM proteins using Ni-nitrilotriacetic acid agarose; (2) cleavage of His-tags using Turbo TEV protease (Accelagen, San Diego, CA, United States); (3) collection of untagged proteins by an additional passage over Ni-nitrilotriacetic acid agarose; (4) SDS-PAGE analysis to assess the purity of the protein products; (5) measurement of protein concentrations by using a Pierce bicinchoninic acid protein assay kit (Thermo Scientific, Waltham, MA, United States); and (6) monitoring β-lactamase activity using nitrocefin (Oxoid, Ltd., Basingstoke, United Kingdom) (Liu et al., 2017).
Enzyme Kinetics Assay
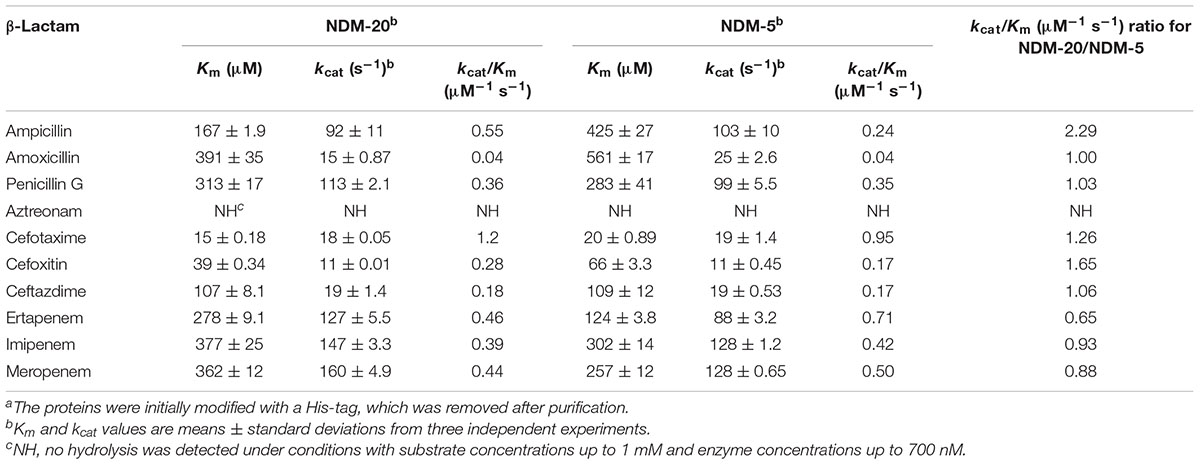
To assay the catalytic properties of NDM-20 and NDM-5, a kinetic study was conducted. A SpectraMax M5 multi-detection microplate reader (Molecular Devices, Sunnyvale, CA, United States) was used to measure initial hydrolysis rates in 30 mM phosphate buffer (30 μM Zn2+, pH 7.0) at 25°C. The hydrolysis rates were then used to determine the Km and kcat values, as well as the kcat/Km ratio, using a Lineweaver–Burk plot. All data used to determine the hydrolysis rates were collected from three individual experiments, and wavelengths and extinction coefficients were applied as previously described (Crowder et al., 1998; Queenan et al., 2010).
Accession Numbers
The sequence of blaNDM-20 gene has been deposited in GenBank under accession no. KY654092 (sequence of blaNDM-20 in Supplementary data 1). The complete nucleotide sequence of plasmid pNDM-20 (sequence of pNDM-20 plasmid in Supplementary data 2) has been deposited as GenBank accession no. MF458176.
Results and Discussion
Strain Features
Escherichia coli strain CCD1 was isolated from a swine fecal swab collected from a commercial pig farm in Shandong province, China. The susceptibility testing results showed that E. coli CCD1 exhibited high level resistance to all β-lactams examined (Table 1), including aztreonam, carbapenems and combinations of penicillins and β-lactamase inhibitors, but susceptible to gentamicin and colistin. The modified Hodge test was positive for carbapenemase, and subsequent positive result from the MBL combined-disk test further suggested MBL production in E. coli CCD1.
The de novo assembled genome of CCD1 showed that the number of contigs was 87 and length of N50 was 110,756 bp. PCR and sequencing analysis identified the presence of a novel NDM-encoding gene, designated blaNDM-20 (GenBank accession no. KY654092). Relative to blaNDM-1, blaNDM-20 contained three point mutations at positions 262 (G→T), 460 (A→C), and 809 (G→A), generating amino acid substitutions Val88Leu, Met154Leu, and Arg270His, respectively (Supplementary Figure S1). Homology and phylogenetic analysis of the amino acid sequences of all NDM variants showed that NDM-20 shared the closest evolutionary relationship with NDM-5 (Figure 1). Notably, only one difference (Arg270His) was observed between the two variants, with this being the first description of a substitution at residue 270. In addition, sequence analysis confirmed the presence of ampC, blaCMY (aztreonam resistance), and sul2 (sulfonamide resistance) in strain CCD1 (Seral et al., 2012). However, no other ESBL-encoding genes were detected.
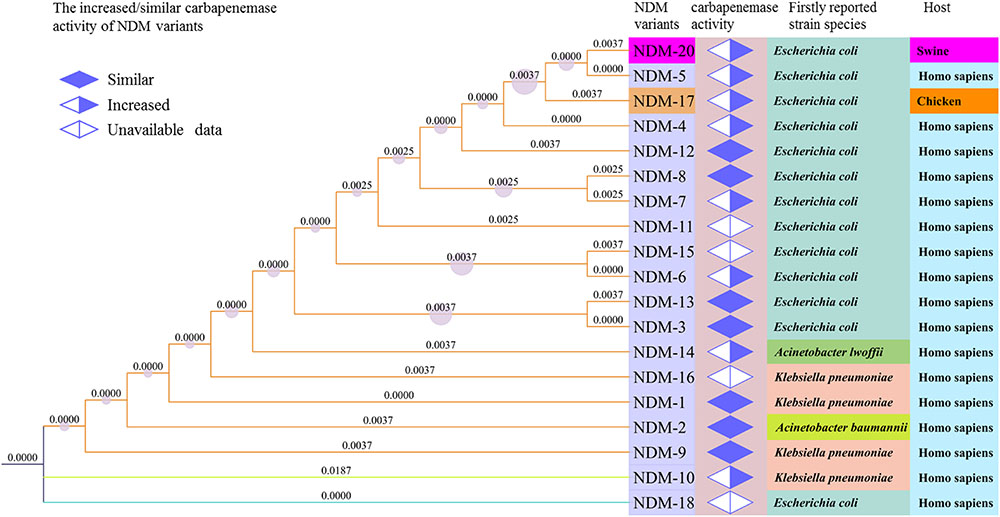
FIGURE 1. Phylogenetic relationships between New Delhi metallo-β-lactamase (NDM) variants based on amino acid sequences. The tree was generated in MEGA7 and iTOL using the maximum likelihood method. Increased/similar carbapenemase activity of NDM variants was determined from previously reported data and compared with NDM-1. The numbers on the branches signify represent branch lengths.
MLST analysis classified E. coli CCD1 as ST1114, suggesting that E. coli ST1114 strains have the potential to become a reservoir for the blaNDM-20-like gene. A previous study showed that ST1114 E. coli isolates are the second largest reservoir of mcr-1 (Matamoros et al., 2017), while our previous study identified two E. coli ST1114 isolates from chickens that were positive for both mcr-1 and blaNDM-5 (Wang et al., 2017).
Transferability of blaNDM-20 and Plasmid Analysis
Conjugation assays confirmed that blaNDM-20 could be transferred between E. coli strains, although the transfer frequency was only 5.01 × 10-9 per donor. Location analysis showed that the blaNDM-20 probe hybridized to a ∼47 kb plasmid band in the transconjugant DNA extracts (Supplementary Figure S2), indicating that blaNDM-20 was located on a transferable plasmid. Subsequently, the entire 46,161-bp blaNDM-20-harboring plasmid, named pNDM-20 (GenBank accession no. MF458176), was sequenced using a WGS approach (Figure 2). pNDM-20 was identified as an IncX3-type plasmid, a major type of plasmid mediating transmission of blaNDM amongst CRE in China (Zhang et al., 2017a). In pNDM-20, blaNDM-20 was flanked in the upstream region by IS3000-ΔISAba125-IS5 and downstream by ble-trpF-dsbC-IS26-ΔumuD, a common blaNDM genetic structure that will be responsible for horizontal transfer of blaNDM-20 among Enterobacteriaceae (Wang et al., 2014; Liu et al., 2017). No resistance genes other than blaNDM-20 and bleMBL were identified on pNDM-20, supporting the observation that the transconjugants only exhibited resistance to β-lactams, and remained sensitive to aztreonam and other classes of antibiotics.
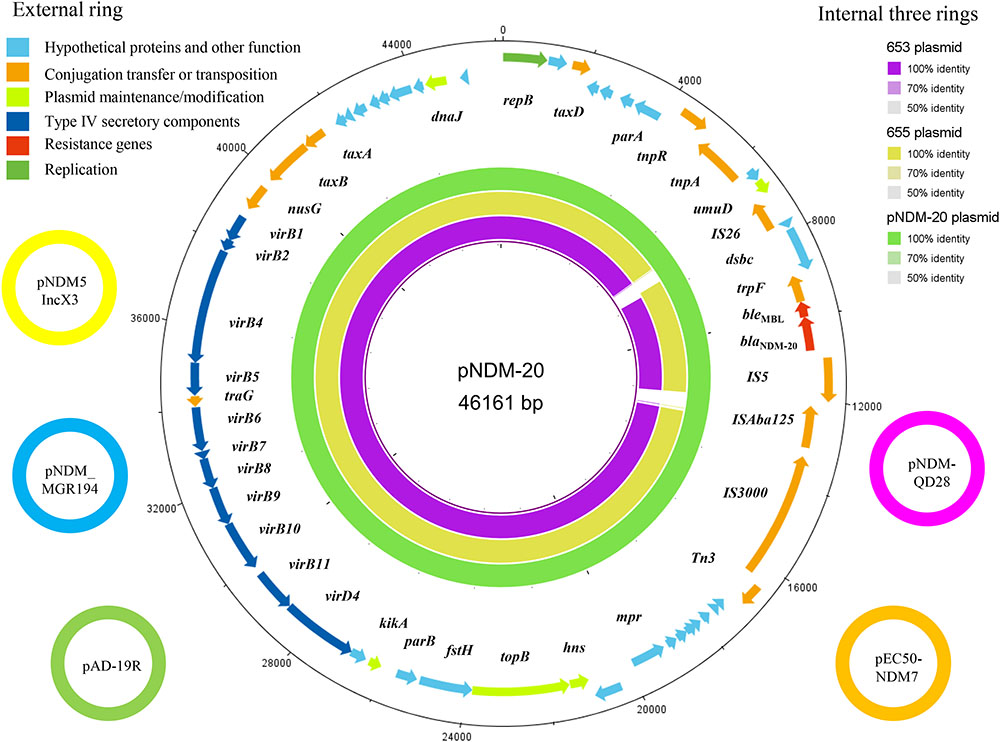
FIGURE 2. The five small external rings represent different blaNDM-harboring plasmids, shown in different colors that had >99% identity. The external ring represents the annotation of plasmid pNDM-20 (GenBank accession no. MF458176). Genes are color-coded depending on functional annotations. The internal three rings represent a comparative analysis of pNDM-20 (green) with two blaNDM-harboring plasmids, 653 (purple) and 655 (yellow) (constructed by BRIG).
Homology analysis revealed 99% nucleotide sequence identity between pNDM-20 and IncX3-type plasmid pNDM-MGR194 (GenBank accession no. KF220657). pNDM-MGR194 was the first reported blaNDM-5-harboring plasmid, and suggested to be responsible for the evolution and distribution of blaNDM-5 amongst bacteria (He et al., 2016). To date, multiple pNDM-MGR194-like plasmids have been identified in bacteria isolated from both humans and animals in China. These plasmids which are 46,161–46,165-bp in length showed >99% nucleotide sequence identity to pNDM-MGR194, including pAD-19R (GenBank accession no. KX833071), pEC50-NDM7 (GenBank accession no. KX470735), pNDM-QD28 (GenBank accession no. KU167608), and pNDM5_IncX3 (GenBank accession no. KU761328) (Figure 2). Therefore, the identification of pNDM-20 provides further evidence that IncX3-type blaNDM-harboring plasmids have become established in bacterial strains originating from both humans and animals, and may circulate via the food chain (He et al., 2016; Liu et al., 2017).
Of note, pNDM-20 had the same backbone structure as the two blaNDM-5-harboring plasmids (653 and 655) from the ST1114 E. coli strains mentioned above. Further analysis showed that the two blaNDM-5-harboring plasmids could be typed as IncX3 using PlasmidFinder software1 (Figure 2). Given these plasmids have the same genetic background and plasmid group/type and were all isolated from ST1114 E. coli strains, it is possible that blaNDM-20 evolved from blaNDM-5 through a series of mutations.
Functional Analysis of blaNDM-20
To determine the specific function of blaNDM-20, blaNDM-20, the reference gene blaNDM-5 (containing only a single nucleotide difference at position 809) and blaNDM-1 were cloned, and the corresponding transformants were examined to determine their antibiotic sensitivity profiles. As shown in Table 1, compared to DH5α+pHSG398, expression of blaNDM-20, blaNDM-5, and blaNDM-1 in E. coli DH5α reduced susceptibility to almost all tested β-lactams, including penicillins, cephalosporins, carbapenems, and combinations of penicillins and β-lactamase inhibitors, except aztreonam. In addition, the MICs of piperacillin was not affected by the addition of β-lactamase inhibitors tazobactam. In general, the blaNDM-20 transformant DH5α+pHSG398/NDM-20 exhibited similar β-lactam resistance to blaNDM-5 transformant DH5α+pHSG398/NDM-5, although several slight differences were observed. Compared with DH5α+pHSG398/NDM-1, the DH5α+pHSG398/NDM-20 exhibited a fourfold elevation in MIC of ertapenem, indicating that blaNDM-20 should enhance resistance to ertapenem. All our findings demonstrated that blaNDM-20 was not as effective as blaNDM-5, but conferred higher resistance to carbapenems than blaNDM-1.
Enzyme Activity Analysis
To further characterize NDM-20 and explore the influence of the amino acid substitutions on enzymatic activity, NDM-20 and NDM-5 were successfully purified (>90%) and their kinetic parameters were examined. NDM-20 was able to hydrolyse all β-lactams tested, except aztreonam (Table 2). The kcat/Km ratios for ampicillin, cefotaxime, and cefoxitin for NDM-20 were higher than those for NDM-5, but carbapenems kcat/Km ratios for NDM-20 were lower than that of NDM-5. These findings resulted from the lower Km values for some penicillins and cephalosporins and higher values for carbapenems for NDM-20 compared with NDM-5. These results demonstrated that NDM-20 had higher enzymatic activity against some penicillins and cephalosporins and lower activity against carbapenems relative to NDM-5, caused by its enhanced affinity for some penicillins and cephalosporins and depressed affinity for carbapenems.
NDM-20 contains an Arg270His substitution relative to NDM-5. This substitution may be responsible for the decreased carbapenemase activity of NDM-20 relative to NDM-5. Two other amino acid substitutions (Val88Leu and Met154Leu) were identified in NDM-20 compared with NDM-1. The Val88Leu substitution was reported to decrease the hydrolytic activity of NDM, while the Met154Leu substitution appeared to increase the carbapenemase activity (Nordmann et al., 2012). The active site of NDM-1 is formed by two loops, loop3 and loop10, and crucial amino acid residues binding to two zinc ions, including His120, His122, His189, His250, Cys208, and Asp124 (Green et al., 2011), and the residue 270 was located out of the enzyme active center (Figure 3). Based on this information, the substitution at residue 270 would not affect the conformation of enzyme active site. It is also possible that the Arg270His substitution may change the protein conformation of NDM-20, thereby altering its hydrolytic activity (Zou et al., 2015). However, further experiments are required to confirm which, if any, of these mechanisms contribute to the altered activity of NDM-20 compared with NDM-5.
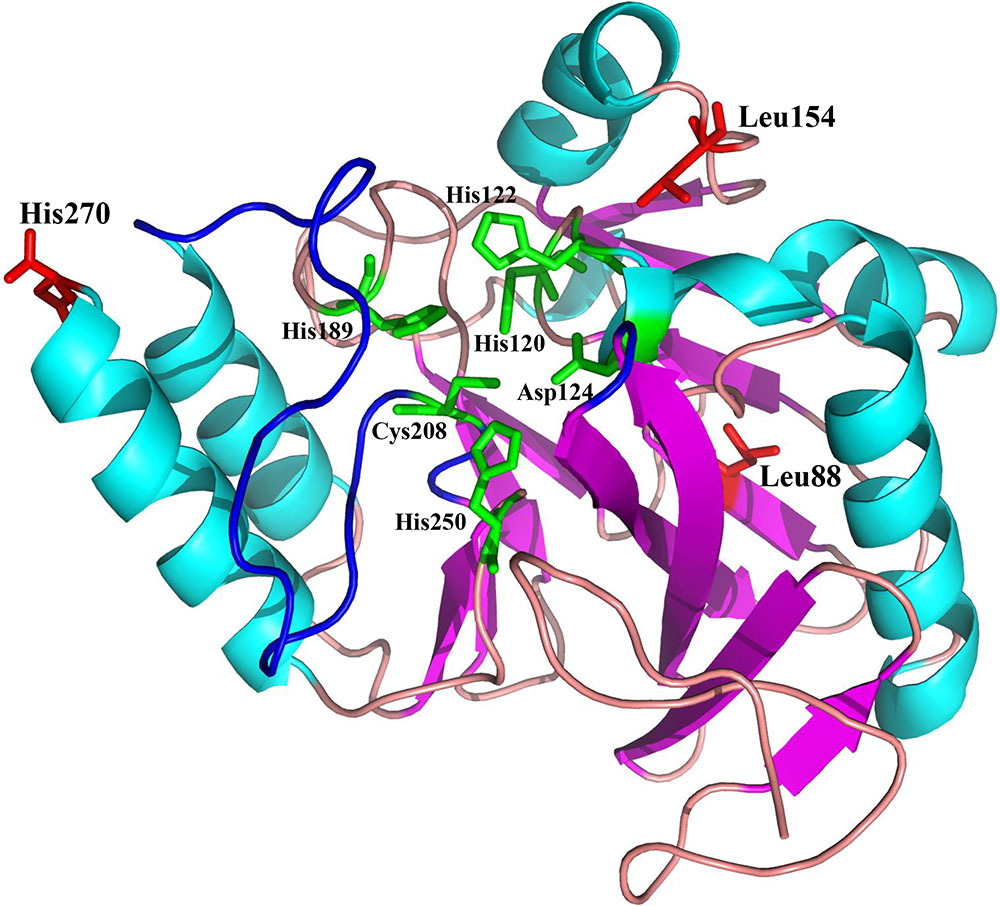
FIGURE 3. Homology model of NDM-20. Protein backbone of NDM-20, shown as a cartoon with the helices and strands. The three amino acid substitutions (L88, L154, and H270) are labeled and colored red.
Conclusion
Our study reports a novel NDM variant, NDM-20, in a ST1114 E. coli isolate recovered from food animal in China. Compared to NDM-5, NDM-20 enhances the hydrolytic activity toward some penicillins and cephalosporins, but depresses the carbapenemase activity, which may affect drug strategies for CRE infection. Both the identification of NDM-20 from swine and NDM-17 from chicken suggest that the food animals have become the reservoir of bacteria producing NDM, which may drive NDM evolution to enhanced enzyme activity toward some or all β-lactams. Furthermore, blaNDM-20 positive bacteria possess the potential capability of dissemination in not only animals but human, becoming a worldwide spread risk. It is time to monitor bacteria producing NDM-20 and other NDM variants in food animals, and coordinate globally to prevent or slow down the NDM evolution.
Author Contributions
JS was responsible for the study design. ZL, JL, XW, and DL assisted in data collection. ZL and YK completed the data interpretation. ZL, YW, and JS wrote the report. All authors revised, reviewed, and approved the final report.
Funding
The study was supported by grants from the National Natural Science Foundation of China (31530076, 31422055, and 81661138002) and the National Basic Research Program of China (2013CB127200).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00248/full#supplementary-material
FIGURE S1 | Secondary structure and disorder prediction of NDM-20. Alignment was constructed in Phyre2 using NDM-1 as template (fold library id: c3rkjA in Phyre). The V88L, M154L, and R270H substitutions are boxed in blue.
FIGURE S2 | Localization of blaNDM-20 on Escherichia coli plasmid pNDM-20 by Southern blot hybridization using a blaNDM-20 probe (A) and by S1-PFGE (B). Lanes 1 and 3, blaNDM-20 transconjugant E. coli 20J3; lanes 2 and 4, E. coli J53; lane M, marker, Salmonella H9812.
DATA 1 | The sequence of bla NDM-20 gene has been deposited in GenBank under accession no. KY654092. The sequence of bla NDM-20 gene as follows:
DATA 2 | The complete nucleotide sequence of plasmid pNDM-20 has been deposited as GenBank accession no. MF458176. The sequence of plasmid pNDM-20 as follows:
TABLE S1 | Amplified and cloning primers.
Footnote
References
Borgia, S., Lastovetska, O., Richardson, D., Eshaghi, A., Xiong, J., Chung, C., et al. (2012). Outbreak of carbapenem-resistant Enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin. Infect. Dis. 55:e109-17. doi: 10.1093/cid/cis737
Clinical and Laboratory Standards Institute [CLSI] (2015). Performance Standards for Antimicrobial Susceptibility Testing; 24th Informational Supplement, M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI).
Crowder, M. W., Walsh, T. R., Banovic, L., Pettit, M., and Spencer, J. (1998). Overexpression, purification, and characterization of the cloned metallo-β-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42, 921–926.
Dallenne, C., Da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
EUCAST (2017). European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org/clinical_breakpoints/
Green, V. L., Verma, A., Owens, R. J., Phillips, S. E. V., and Carr, S. B. (2011). Structure of New Delhi metallo-β-lactamase 1 (NDM-1). Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1160–1164. doi: 10.1107/S1744309111029654
He, T., Wang, Y., Sun, L., Pang, M., Zhang, L., and Wang, R. (2016). Occurrence and characterization of blaNDM-5-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu. China. J. Antimicrob. Chemother. 72, 90–94. doi: 10.1093/jac/dkw357
Inouye, M., Dashnow, H., Raven, L. A., Schultz, M. B., Pope, B. J., Tomita, T., et al. (2014). SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6:90 doi: 10.1186/s13073-014-0090-6
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 17:101. doi: 10.1186/s12866-017-1012-8
Liu, Z., Wang, Y., Walsh, T. R., Liu, D., Shen, Z., Zhang, R., et al. (2017). Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob. Agents Chemother. 61:e02233-16. doi: 10.1128/AAC.02233-16
Madec, J., Haenni, M., Nordmann, P., and Poirel, L. (2017). Extended-spectrum β-lactamase/AmpC-and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans. Clin. Microbiol. Infect. 23, 826–833. doi: 10.1016/j.cmi.2017.01.013
Matamoros, S., van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
Nordmann, P., Boulanger, A. E., and Poirel, L. (2012). NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 56, 2184–2186. doi: 10.1128/AAC.05961-11
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Queenan, A. M., Shang, W., Flamm, R., and Bush, K. (2010). Hydrolysis and inhibition profiles of β-lactamases from molecular classes A to D with doripenem, imipenem, and meropenem. Antimicrob. Agents Chemother. 54, 565–569. doi: 10.1128/AAC.01004-09
Seral, C., Gude, M. J., and Castillo, F. J. (2012). Emergence of plasmid mediated AmpC β-lactamasas: origin, importance, detection and therapeutical options. Rev. Esp. Quimioter. 25, 89–99.
Wang, X., Xu, X., Li, Z., Chen, H., Wang, Q., Yang, P., et al. (2014). An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb. Drug Resist. 20, 144–149. doi: 10.1089/mdr.2013.0100
Wang, Y., He, T., Schwarz, S., Zhao, Q., Shen, Z., Wu, C., et al. (2013). Multidrug resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int. J. Med. Microbiol. 303, 84–87. doi: 10.1016/j.ijmm.2012.12.004
Wang, Y., Zhang, R., Li, J., Wu, Z., Yin, W., Schwarz, S., et al. (2017). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2:16260. doi: 10.1038/nmicrobiol.2016.260
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017a). Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Zhang, R., Liu, Z., Li, J., Lei, L., Yin, W., Li, M., et al. (2017b). Presence of VIM positive Pseudomonas species in chickens and their surrounding environment. Antimicrob. Agents Chemother. 61:e00167-17. doi: 10.1128/AAC.00167-17
Keywords: NDM-20, carbapenemase, ST1114, IncX3, kinetic parameters
Citation: Liu Z, Li J, Wang X, Liu D, Ke Y, Wang Y and Shen J (2018) Novel Variant of New Delhi Metallo-β-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 9:248. doi: 10.3389/fmicb.2018.00248
Received: 04 November 2017; Accepted: 31 January 2018;
Published: 21 February 2018.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Benjamin Andrew Evans, University of East Anglia, United KingdomTommaso Giani, University of Siena, Italy
Copyright © 2018 Liu, Li, Wang, Liu, Ke, Wang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianzhong Shen, sjz@cau.edu.cn
 Zhihai Liu
Zhihai Liu Jiyun Li
Jiyun Li Xiaoming Wang1
Xiaoming Wang1 Yang Wang
Yang Wang Jianzhong Shen
Jianzhong Shen