- Section of Virology, Division of Infectious Diseases, Department of Medicine, Imperial College London, London, United Kingdom
HTLV-1 plus-strand transcription begins with the production of doubly-spliced tax/rex transcripts, the levels of which are usually undetectable in freshly isolated peripheral blood mononuclear cells (PBMCs) from HTLV-1-infected individuals. However, the presence of a sustained chronically active cytotoxic T-cell response to HTLV-1 antigens in virtually all HTLV-1-infected individuals, regardless of their proviral load, argues against complete latency of the virus in vivo. There is an immediate burst of plus-strand transcription when blood from infected individuals is cultured ex vivo. How is the HTLV-1 plus strand silenced in PBMCs? Is it silenced in other anatomical compartments within the host? What reactivates the latent provirus in fresh PBMCs? While plus-strand transcription of the provirus appears to be intermittent, the minus-strand hbz transcripts are present in a majority of cells, albeit at low levels. What regulates the difference between the 5′- and 3′-LTR promoter activities and thereby the tax-hbz interplay? Finally, T lymphocytes are a migratory population of cells that encounter variable environments in different compartments of the body. Could these micro-environment changes influence the reactivation kinetics of the provirus? In this review we discuss the questions raised above, focusing on the early events leading to HTLV-1 reactivation from latency, and suggest future research directions.
Introduction
Human T-lymphotropic virus type 1 (HTLV-1), also known as Human T-cell leukemia virus type 1, is a retrovirus that mainly infects CD4+ T-cells in vivo. In ∼5–10% of the infected individuals, HTLV-1 infection leads to either an aggressive T-cell malignancy, adult T-cell leukemia/lymphoma (ATL), or a chronic progressive neuro-inflammatory condition called HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). Both these conditions have limited treatment options, and ATL in particular carries a very poor prognosis.
Initial infection is asymptomatic and occurs through breast milk, blood, or semen of infected individuals (Bangham and Matsuoka, 2017). HTLV-1 replicates in the host through two distinct routes: (i) Infectious spread: this mode of spread involves productive replication from the integrated provirus followed by the transfer of newly produced virions through the virological synapse (Igakura et al., 2003; Pais-Correia et al., 2010). This is the major route of viral spread in the initial stages of infection when the proviral load – the percentage of infected PBMCs – is low (Bangham et al., 2014). Infectious spread results in the formation of distinct T-cell clones, each clone carrying a single-copy HTLV-1 provirus integrated in a unique genomic location within the host genome. (ii) Mitotic spread: proliferation of HTLV-1-infected host cells results in passive replication of the integrated HTLV-1 provirus within their genome. The two daughter cells resulting from mitosis of an HTLV-1-infected parent cell carry the provirus in the same genomic integration site. In contrast to infectious spread, the contribution of this mode to the proviral load in infected individuals may be small in the early stages of infection but gradually increases during the chronic stage of infection (Bangham et al., 2014). Although the proviral load in each host can fluctuate by a small factor (2- to 5-fold) over time, the proviral loads can vary between infected individuals by over 1000-fold (Nagai et al., 1998; Demontis et al., 2013). Individuals with a higher proviral load are at greater risk of developing either ATL or HAM/TSP (Matsuzaki et al., 2001; Iwanaga et al., 2010). An infected individual typically carries about 104 to 105 different T-cell clones, each with a unique proviral integration site (Bangham et al., 2014). The extent of proliferation of HTLV-1-infected T-cell clones, and thus their respective contribution to an individual’s proviral load, both vary greatly from one clone to another.
A peculiar characteristic of HTLV-1 is the absence of detectable cell-free virions in infected individuals (Demontis et al., 2015). HTLV-1 was previously considered to be latent in infected individuals because there are no detectable plus-strand viral structural RNA or protein products in the peripheral blood mononuclear cells (PBMCs) freshly obtained from HTLV-1 infected individuals. Also, HTLV-1 is genetically stable, with minimal sequence variation over evolutionary time (Gessain et al., 1992), suggesting that de novo viral replication, which generates sequence variation, contributes little to the long-term persistence of HTLV-1 in vivo. However, a sustained chronically active CTL response to HTLV-1 can be observed in virtually all infected individuals (Jacobson et al., 1990; Bangham, 2009). This observation suggests that the host immune system routinely encounters viral antigens in vivo. Since the HTLV-1 plus-strand appears to be latent in peripheral blood, the implication is that plus-strand expression occurs either in intermittent bursts, or outside the circulation, or both. Thus it is necessary to understand the determinants of HTLV-1 latency and reactivation in vivo (Figure 1) to devise effective preventive and therapeutic approaches against HTLV-1-associated diseases such as HAM/TSP and ATL.
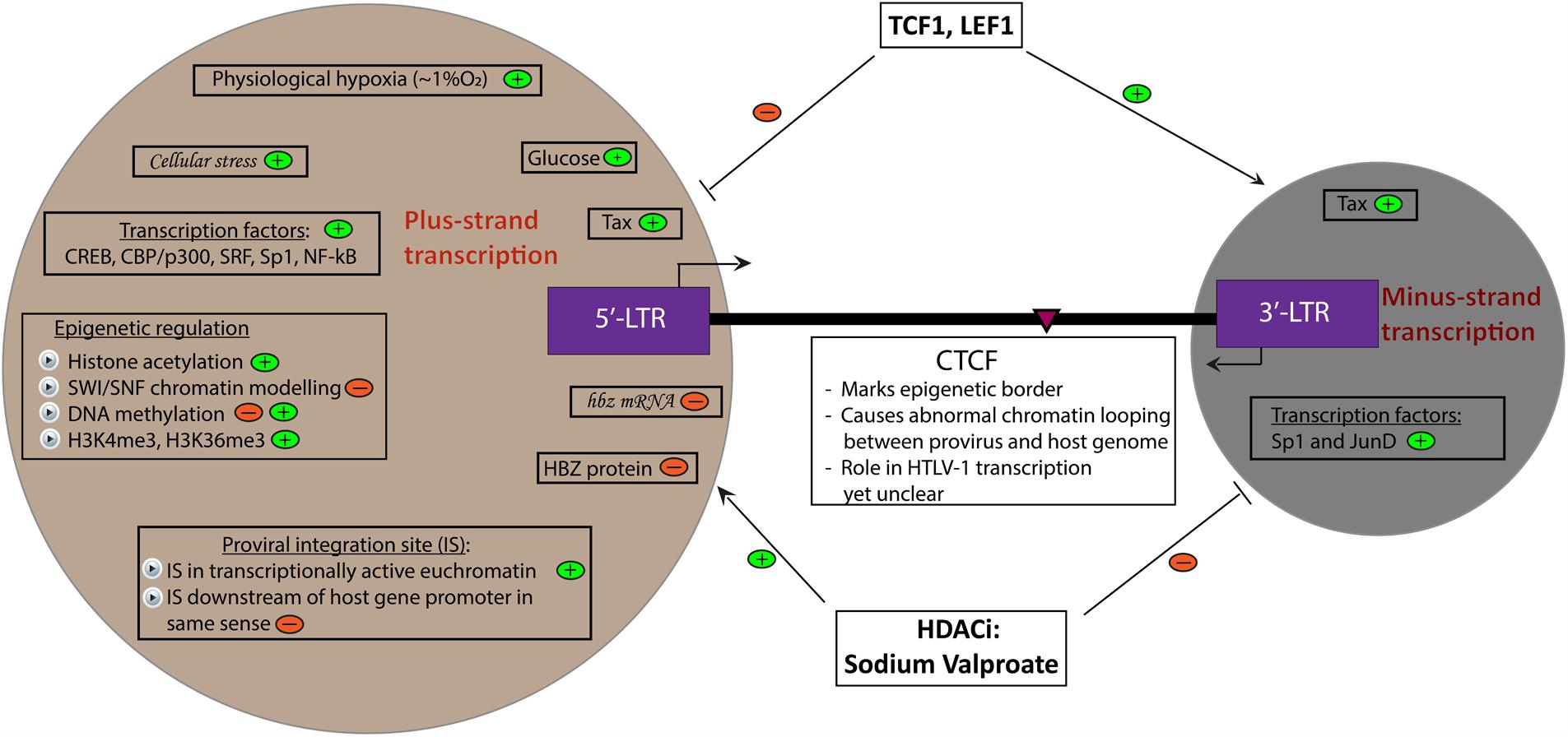
FIGURE 1. Determinants of HTLV-1 plus- and minus-strand transcription. A summary of currently known factors that induce (depicted by  ) and inhibit (depicted by
) and inhibit (depicted by  ) plus- and minus-strand transcription of the HTLV-1 provirus. Italicized words denote hypothetical factors. The pink arrowhead marks the position of the CTCF binding site in the provirus.
) plus- and minus-strand transcription of the HTLV-1 provirus. Italicized words denote hypothetical factors. The pink arrowhead marks the position of the CTCF binding site in the provirus.
Proviral Genomic Integration Site
HTLV-1 cannot be looked at in isolation because it is an inseparable part of the chromatinized host genomic DNA. Can we predict the behavior of the integrated provirus in the context of its flanking host genome? HTLV-1 integration favors transcriptionally active regions of the host genome (Melamed et al., 2013). Recently, the host enzyme Protein Phosphatase 2A (PP2A) was identified as a major host co-factor for the HTLV-1 integrase, which could influence the selection of genomic integration sites (Maertens, 2016). Furthermore, plus-strand transcription is silenced when the viral DNA is integrated downstream of a host gene promoter in the same-sense orientation, possibly by transcriptional interference (Melamed et al., 2013). Similarly, the presence of the SWI/SNF-associated ATPase BRG-1 (identified by chromatin immunoprecipitation) upstream of the integrated HTLV-1 provirus is associated with silencing of plus-strand transcription, but its presence downstream of the HTLV-1 sequence is associated with proviral plus-strand expression (Melamed et al., 2013). Recently, it was reported that integration of HTLV-1 in the vicinity of cancer driver genes causes either premature interruption of transcription or antisense-dependent cis-perturbation of these genes, perhaps contributing to leukemogenesis (Rosewick et al., 2017).
The discovery of a functional CTCF binding site within the pX region of the HTLV-1 genome adds a new dimension to the importance of the flanking genome to proviral transcription (Satou et al., 2016). CTCF is a critical host protein which binds to an insulator motif in the DNA and appears to limit the spread of epigenetic modifications (Merkenschlager and Nora, 2016). Consistent with this function, the CTCF binding region in HTLV-1 is associated with the presence of a sharp epigenetic border in several histone and DNA modifications. It is possible that the epigenetic border formed by CTCF binding within the HTLV-1 provirus allows unhindered minus-strand transcription while reversibly inhibiting the plus-strand activity. Another central function of CTCF is the formation of chromatin loops, which regulate contacts between enhancers and promoters. It is thus possible that the CTCF bound to the provirus produces abnormal chromatin loops, resulting in aberrant cellular gene expression (Cook et al., 2017). Experiments are now in progress to test these hypotheses. Ultimately, this aberrant gene expression may be an important factor in the development of ATL.
The Tax-HBZ Interplay
Gene transcription in HTLV-1 progresses in both the sense and anti-sense directions, governed by promoters in the identical long terminal repeats (LTRs) situated at each end of the genome.
Plus-Strand Transcription
This is regulated by the TATA-box-containing inducible promoter in the 5′-LTR (Miyazato et al., 2016). TATA boxes are frequently observed in promoters of cellular genes which are highly regulated and responsive to stress and extracellular signals (Basehoar et al., 2004; Bahrami and Drablos, 2016). The 5′-LTR contains three tandem 21-bp imperfect nucleotide repeats called tax-responsive elements (TREs) which bind to the ATF/CREB family of proteins to activate HTLV-1 plus-strand transcription (Armstrong et al., 1993). The transactivator protein Tax, encoded by the pX region of the HTLV-1 genome by a double-splicing mechanism, is the most potent activator of plus-strand transcription and initiates a positive-feedback loop which induces transcription and promotes replication (Beimling and Moelling, 1992; Anderson and Dynan, 1994). Tax enhances the binding of CREB to the TRE, thereby promoting the transactivation of the proviral plus-strand (Suzuki et al., 1993). In addition to ATF/CREB family, the 5′-LTR also has binding sites for several major transcription factors, namely NF-κB, SRF and Sp1, which play direct or indirect roles in activating plus-strand transcription (Armstrong et al., 1993). In addition to Tax, the HTLV-1 pX region encodes another regulatory protein, Rex, which regulates the stabilization of unspliced and singly spliced viral mRNAs, their nuclear export and subsequent effective translation of the viral proteins (Nakano and Watanabe, 2012). The other HTLV-1 pX region plus-strand products play accessory roles in vivo: p30 and p13 promote infectivity in a rabbit model (Bartoe et al., 2000; Silverman et al., 2004; Hiraragi et al., 2006), and p30 and p12/p8 enhance persistent infection in macaques (Valeri et al., 2010; Pise-Masison et al., 2014). p13 increases the production of reactive oxygen species, selectively killing transformed cells, and may favor persistence in vivo (Silic-Benussi et al., 2010). However, these genes are not essential for viral replication or T-cell immortalization (Derse et al., 1997; Robek et al., 1998), and may not be required for transmission (Furukawa et al., 2004).
Minus-Strand Transcription
This is initiated at the 3′-end of the provirus and progresses in the anti-sense direction (Gaudray et al., 2002). It is governed by the TATA-less promoter in the 3′-LTR (Yoshida et al., 2008), which contains three TREs and three Sp1 binding sites (Yoshida et al., 2008; Ma et al., 2016). sHBZ is the major transcript driven by the 3′-LTR.
The interplay between the plus-strand and minus-strand transcription is a critical and poorly understood aspect of the HTLV-1 life cycle. We infer that Tax expression in vivo is intermittent and highly regulated as it is a very immunogenic protein (Jacobson et al., 1990; Parker et al., 1992). There is no detectable plus-strand gene expression in freshly isolated PBMCs from HTLV-1 infected individuals (Kinoshita et al., 1989; Rende et al., 2011). HBZ, in contrast, is poorly immunogenic and is expressed at a very low level; however, the presence of a detectable cytotoxic T-lymphocyte (CTL) response to HBZ is associated with a lower proviral load (MacNamara et al., 2010). This constant, low-level hbz mRNA expression is thought to be necessary for the persistence of HTLV-1 in vivo: hbz expression can be observed in all HTLV-1-infected individuals, including those with HAM/TSP and ATL (Yoshida et al., 2008). Tax and HBZ have strong effects on a number of cellular processes and signaling cascades (Ma et al., 2016); the effects of the two proteins frequently act in opposition. Certain transcription factors including TCF1 and LEF1 inhibit plus-strand transcription from the 5′-LTR while simultaneously enhancing minus-strand transcription from the 3′-LTR (Ma et al., 2015). In contrast, the histone deacetylase inhibitor (HDACi) valproate activates plus-strand transcription while inhibiting expression of the minus-strand (Lezin et al., 2007; Belrose et al., 2011; Olindo et al., 2011). It is also known that Tax activates minus-strand transcription through binding of the TREs in the 3′-LTR (Landry et al., 2009). Recently, it was shown by single-molecule RNA-FISH that, at a single-cell level, both plus- and minus-strand transcription occurs in bursts, albeit at different times and intensities. HTLV-1 plus-strand mRNA transcription is enhanced in the absence of minus-strand transcripts, while high-level plus-strand transcription promotes transcription from the minus-strand (Billman et al., 2017), generating a putative negative feedback inhibition of plus-strand expression. Until now, it was believed that all HTLV-1-infected cells express hbz mRNA at all times; however, this study showed that, in naturally-infected T-cell clones isolated by limiting dilution from HTLV-1-infected individuals, not all HTLV-1-infected cells in each clone are hbz positive (Billman et al., 2017) at a given time. Many questions remain unanswered. Most hbz transcripts were found in the nuclei of cells, consistent with previous observations (Rende et al., 2011) and with the low level of protein expression of HBZ (Shiohama et al., 2016). HBZ protein inhibits tax expression by competing for binding of the transcription co-factor CREB (Gaudray et al., 2002; Lemasson et al., 2007; Clerc et al., 2008). However, since HBZ protein is expressed at such a low level in naturally-infected cells, the importance of this mechanism is uncertain (Shiohama et al., 2016).
These observations raise important questions: does hbz RNA also inhibit tax expression? What are the factors that regulate minus-strand transcriptional bursts? It has been shown that Sp1 co-operates with JunD to activate minus-strand transcription, but how and when this happens in the context of the cell cycle and viral replication are still unknown (Gazon et al., 2012). Recently, Billman et al. (2017) showed at the single cell level, in naturally-infected HTLV-1 positive T-cell clones, that cells in S and G2/M phase of the cell cycle have elevated tax and hbz mRNA levels, suggesting that the cell cycle plays a role in regulating HTLV-1 transcription. It is now becoming clear that the results of population-level analyses can mask strong single-cell heterogeneity within the population.
Epigenetic Regulation of HTLV-1 Transcription
Small-molecule inhibitors targeting epigenetic enzymes have been widely used in HIV latency research. Indeed, reactivation of the latent HIV-1 provirus occurs in response to HDACi [e.g., suberanilohydroxamic acid (SAHA)], DNA methylation inhibitors (e.g., 5-azacytidine) and histone methyltransferase inhibitors (e.g., the EZH1/2 inhibitor GSK343) (Spina et al., 2013; Tripathy et al., 2015). Similarly, in the case of HTLV-1, the HDACi valproate and DNA methyltransferase inhibitor 5-azacytidine have been shown to activate the latent HTLV-1 provirus (Koiwa et al., 2002; Belrose et al., 2011). These observations suggest that epigenetic modulations in the promoters, enhancers and gene bodies of the integrated provirus play an important role in proviral transcription. Cytosine methylation of the 5′-LTR of HTLV-1 is associated with silencing of plus-strand transcription; the degree of this 5′-LTR methylation varies from individual to individual, whereas the 3′-LTR is unmethylated in most individuals (Taniguchi et al., 2005), consistent with the constitutive activation of the promoter in the 3′-LTR. However, it is unclear how dynamic the DNA methylation pattern of the provirus is. It is unlikely that the highly regulated promoter in the 5′-LTR of HTLV-1 would rely primarily on DNA demethylation for its reactivation. Data are needed on the extent of DNA methylation during bursts of gene expression from the plus-strand and minus-strand, and whether this methylation is a cause or an effect of promoter activation. Activation of plus-strand transcription is associated with Tax-mediated recruitment of pCREB and CBP/p300, resulting in histone acetylation and nucleosomal remodeling through BRG-1-containing SWI/SNF complexes (Sharma and Nyborg, 2008; Easley et al., 2010). Similarly, HTLV-1 plus-strand transcription is associated with a significant increase in the activatory histone epigenetic modifications H3K4me3 and H3K36me3 in different regions of the HTLV-1 provirus (Kulkarni et al., 2017). It is likely that, as in host gene expression, many epigenetic modifications are a cause as well as consequence of viral reactivation. Most of the epigenetic studies carried out so far have been in in vitro cellular models where Tax expression is the primary mechanism of 5′-LTR activation through a positive feedback loop. However, evidence from freshly isolated PBMCs from HTLV-1-infected individuals indicates that there is little or no Tax protein expression in vivo at steady state. Hence, it is important to examine the earliest epigenetic changes that occur at the provirus in freshly isolated PBMCs from HTLV-1-infected individuals to provide clues to the changes that occur in vivo. It is still unclear what primarily reactivates plus-strand transcription in ex vivo PBMCs from HTLV-1-infected individuals. The answer to this conundrum may elucidate the key question – what regulates the rapidly reversible silencing of plus-strand transcription in vivo?
Metabolic Regulation of HTLV-1 Transcription
The metabolic and functional activities of circulating PBMCs depend directly on their micro-environment. What effects do metabolic changes have on the integrated HTLV-1 provirus within these cells? We recently showed that HTLV-1 reactivation from latency is severely limited in the absence of glucose in the surrounding medium (Kulkarni et al., 2017). In support of this conclusion, we found that PBMCs treated with glycolysis inhibitors – and to a lesser extent with mitochondrial electron transport chain (ETC) inhibitors – show a significant impairment in HTLV-1 plus-strand transcription when they are cultured ex vivo. The importance of glucose in the HTLV-1 life cycle is emphasized by the fact that the glucose receptor GLUT-1 is a cellular receptor for HTLV-1 (Manel et al., 2003). Surprisingly, drugs that inhibit or stimulate the TCA cycle do not alter HTLV-1 transcription (Kulkarni et al., 2017). Indeed, it is known that different subsets of T-cells use distinct metabolic pathways to satisfy their energy requirements (Dimeloe et al., 2017). How glucose metabolism influences HTLV-1 reactivation from latency is not known. A plausible explanation is that inhibition of glycolysis or the mitochondrial ETC reduces the availability of Acetyl CoA and ATP. Acetyl CoA is an important acetyl group donor in the process of histone acetylation, which is associated with transcriptional activation. Similarly, the SWI/SNF family of chromatin-remodeling complexes such as BAF and PBAF rely on energy in the form of ATP for their nucleosomal remodeling function. It has been shown before that HTLV-1 plus-strand activation is intimately linked with histone acetylation and chromatin remodeling at the 5′-LTR promoter (Easley et al., 2010; Nyborg et al., 2010). Another important question that arises from these results is the possible impact of high blood glucose levels on HTLV-1 replication. Do HTLV-1-infected individuals with poorly controlled diabetes have a higher proviral load of HTLV-1, resulting in a significantly higher risk of HTLV-1-associated diseases such as HAM/TSP or ATL? Further work is needed to clarify the cause and effect relationships between metabolic activity and proviral activation in HTLV-1-infected primary CD4+ T-cells.
Anatomical Regulation of HTLV-1 Transcription
Lymphocytes are constantly exposed to highly variable stresses during circulation through different body compartments. As mentioned above, HTLV-1 appears to be latent in freshly isolated PBMCs from the peripheral blood of HTLV-1 infected individuals. The presence of a chronically active CTL response to HTLV-1 in all infected individuals raises the possibility that HTLV-1 expression is more frequent in different compartments like the bone marrow, lymph, and lymph nodes (Bangham, 2008). Also, Yasunaga et al. (2016) found that tax transcription in the bone marrow of STLV-1-infected Japanese macaques was significantly higher than that in other tissues. One factor that differs significantly between these compartments is the oxygen tension. While the lymph, lymph nodes, and bone marrow are highly hypoxic (∼1% oxygen), PBMCs in peripheral venous blood are exposed to significantly higher levels of oxygen (∼6–10%) (Tsai et al., 2004; Hangai-Hoger et al., 2007). We recently reported that, in primary PBMCs isolated from HTLV-1 infected individuals, physiological hypoxia (∼1% oxygen) enhances plus-strand HTLV-1 transcription (Kulkarni et al., 2017). Again, further work is needed to identify the molecular mechanism of this effect, and the consequences of this observation on the establishment and spread of viral infection within and between individuals.
Summary
The transcriptional regulation of the plus- and minus-strands of HTLV-1 is affected by a number of virus-specific and host-specific factors. Advances in single-cell genomics and transcriptomics have made it possible to study HTLV-1 infection at the single-cell level. We anticipate that these approaches will shed light on the effect of cellular heterogeneity on viral replication and help identify novel mechanisms that were missed by earlier studies which depended on population-level analyses. Precise identification of the molecular mechanisms involved in HTLV-1 reactivation from latency would help us design and test novel therapeutic strategies to reduce the disease burden associated with this infection.
Author Contributions
Both authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by a Project Grant from the Medical Research Council, United Kingdom (MR/K019090/1), a Senior Investigator award from the Wellcome Trust, United Kingdom (WT100291MA) to CB, and the Imperial National Institute for Health Research Biomedical Research Centre (https://imperialbrc.nihr.ac.uk/).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jocelyn Turpin and Goedele Maertens for comments on the manuscript.
References
Anderson, M. G., and Dynan, W. S. (1994). Quantitative studies of the effect of HTLV-I tax protein on CREB protein-DNA binding. Nucleic Acids Res. 22, 3194–3201. doi: 10.1093/nar/22.15.3194
Armstrong, A. P., Franklin, A. A., Uittenbogaard, M. N., Giebler, H. A., and Nyborg, J. K. (1993). Pleiotropic effect of the human T-cell leukemia virus Tax protein on the DNA binding activity of eukaryotic transcription factors. Proc. Natl. Acad. Sci. U.S.A. 90, 7303–7307. doi: 10.1073/pnas.90.15.7303
Bahrami, S., and Drablos, F. (2016). Gene regulation in the immediate-early response process. Adv. Biol. Regul. 62, 37–49. doi: 10.1016/j.jbior.2016.05.001
Bangham, C. R. (2008). HTLV-1 infection: role of CTL efficiency. Blood 112, 2176–2177. doi: 10.1182/blood-2008-06-163071
Bangham, C. R. (2009). CTL quality and the control of human retroviral infections. Eur. J. Immunol. 39, 1700–1712. doi: 10.1002/eji.200939451
Bangham, C. R., Cook, L. B., and Melamed, A. (2014). HTLV-1 clonality in adult T-cell leukaemia and non-malignant HTLV-1 infection. Semin. Cancer Biol. 26, 89–98. doi: 10.1016/j.semcancer.2013.11.003
Bangham, C. R. M., and Matsuoka, M. (2017). Human T-cell leukaemia virus type 1: parasitism and pathogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160272. doi: 10.1098/rstb.2016.0272
Bartoe, J. T., Albrecht, B., Collins, N. D., Robek, M. D., Ratner, L., Green, P. L., et al. (2000). Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J. Virol. 74, 1094–1100. doi: 10.1128/JVI.74.3.1094-1100.2000
Basehoar, A. D., Zanton, S. J., and Pugh, B. F. (2004). Identification and distinct regulation of yeast TATA box-containing genes. Cell 116, 699–709. doi: 10.1016/S0092-8674(04)00205-3
Beimling, P., and Moelling, K. (1992). Direct interaction of CREB protein with 21 bp Tax-response elements of HTLV-ILTR. Oncogene 7, 257–262.
Belrose, G., Gross, A., Olindo, S., Lezin, A., Dueymes, M., Komla-Soukha, I., et al. (2011). Effects of valproate on Tax and HBZ expression in HTLV-1 and HAM/TSP T lymphocytes. Blood 118, 2483–2491. doi: 10.1182/blood-2010-11-321364
Billman, M. R., Rueda, D., and Bangham, C. R. M. (2017). Single-cell heterogeneity and cell-cycle-related viral gene bursts in the human leukaemia virus HTLV-1. Wellcome Open Res. 2:87. doi: 10.12688/wellcomeopenres.12469.2
Clerc, I., Polakowski, N., Andre-Arpin, C., Cook, P., Barbeau, B., Mesnard, J. M., et al. (2008). An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J. Biol. Chem. 283, 23903–23913. doi: 10.1074/jbc.M803116200
Cook, L., Melamed, A., Yaguchi, H., and Bangham, C. R. (2017). The impact of HTLV-1 on the cellular genome. Curr. Opin. Virol. 26, 125–131. doi: 10.1016/j.coviro.2017.07.013
Demontis, M. A., Hilburn, S., and Taylor, G. P. (2013). Human T cell lymphotropic virus type 1 viral load variability and long-term trends in asymptomatic carriers and in patients with human T cell lymphotropic virus type 1-related diseases. AIDS Res. Hum. Retroviruses 29, 359–364. doi: 10.1089/AID.2012.0132
Demontis, M. A., Sadiq, M. T., Golz, S., and Taylor, G. P. (2015). HTLV-1 viral RNA is detected rarely in plasma of HTLV-1 infected subjects. J. Med. Virol. 87, 2130–2134. doi: 10.1002/jmv.24264
Derse, D., Mikovits, J., and Ruscetti, F. (1997). X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology 237, 123–128. doi: 10.1006/viro.1997.8781
Dimeloe, S., Burgener, A. V., Grahlert, J., and Hess, C. (2017). T-cell metabolism governing activation, proliferation and differentiation; a modular view. Immunology 150, 35–44. doi: 10.1111/imm.12655
Easley, R., Carpio, L., Guendel, I., Klase, Z., Choi, S., Kehn-Hall, K., et al. (2010). Human T-lymphotropic virus type 1 transcription and chromatin-remodeling complexes. J. Virol. 84, 4755–4768. doi: 10.1128/JVI.00851-09
Furukawa, Y., Usuku, K., Izumo, S., and Osame, M. (2004). Human T cell lymphotropic virus type I (HTLV-I) p12I is dispensable for HTLV-I transmission and maintenance of infection in vivo. AIDS Res. Hum. Retroviruses 20, 1092–1099. doi: 10.1089/aid.2004.20.1092
Gaudray, G., Gachon, F., Basbous, J., Biard-Piechaczyk, M., Devaux, C., and Mesnard, J. M. (2002). The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76, 12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002
Gazon, H., Lemasson, I., Polakowski, N., Cesaire, R., Matsuoka, M., Barbeau, B., et al. (2012). Human T-cell leukemia virus type 1 (HTLV-1) bZIP factor requires cellular transcription factor JunD to upregulate HTLV-1 antisense transcription from the 3’ long terminal repeat. J. Virol. 86, 9070–9078. doi: 10.1128/JVI.00661-12
Gessain, A., Gallo, R. C., and Franchini, G. (1992). Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J. Virol. 66, 2288–2295.
Hangai-Hoger, N., Tsai, A. G., Cabrales, P., and Intaglietta, M. (2007). Terminal lymphatics: the potential “lethal corner” in the distribution of tissue pO2. Lymphat. Res. Biol. 5, 159–168. doi: 10.1089/lrb.2007.5303
Hiraragi, H., Kim, S. J., Phipps, A. J., Silic-Benussi, M., Ciminale, V., Ratner, L., et al. (2006). Human T-lymphotropic virus type 1 mitochondrion-localizing protein p13(II) is required for viral infectivity in vivo. J. Virol. 80, 3469–3476. doi: 10.1128/JVI.80.7.3469-3476.2006
Igakura, T., Stinchcombe, J. C., Goon, P. K., Taylor, G. P., Weber, J. N., Griffiths, G. M., et al. (2003). Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science 299, 1713–1716. doi: 10.1126/science.1080115
Iwanaga, M., Watanabe, T., Utsunomiya, A., Okayama, A., Uchimaru, K., Koh, K. R., et al. (2010). Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: a nationwide prospective study in Japan. Blood 116, 1211–1219. doi: 10.1182/blood-2009-12-257410
Jacobson, S., Shida, H., Mcfarlin, D. E., Fauci, A. S., and Koenig, S. (1990). Circulating CD8+ cytotoxic T lymphocytes specific for HTLV-I pX in patients with HTLV-I associated neurological disease. Nature 348, 245–248. doi: 10.1038/348245a0
Kinoshita, T., Shimoyama, M., Tobinai, K., Ito, M., Ito, S., Ikeda, S., et al. (1989). Detection of mRNA for the tax1/rex1 gene of human T-cell leukemia virus type I in fresh peripheral blood mononuclear cells of adult T-cell leukemia patients and viral carriers by using the polymerase chain reaction. Proc. Natl. Acad. Sci. U.S.A. 86, 5620–5624. doi: 10.1073/pnas.86.14.5620
Koiwa, T., Hamano-Usami, A., Ishida, T., Okayama, A., Yamaguchi, K., Kamihira, S., et al. (2002). 5’-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 76, 9389–9397. doi: 10.1128/JVI.76.18.9389-9397.2002
Kulkarni, A., Mateus, M., Thinnes, C. C., Mccullagh, J. S., Schofield, C. J., Taylor, G. P., et al. (2017). Glucose metabolism and oxygen availability govern reactivation of the latent human retrovirus HTLV-1. Cell Chem. Biol. 24, 1377.e3–1387.e3. doi: 10.1016/j.chembiol.2017.08.016
Landry, S., Halin, M., Vargas, A., Lemasson, I., Mesnard, J. M., and Barbeau, B. (2009). Upregulation of human T-cell leukemia virus type 1 antisense transcription by the viral tax protein. J. Virol. 83, 2048–2054. doi: 10.1128/JVI.01264-08
Lemasson, I., Lewis, M. R., Polakowski, N., Hivin, P., Cavanagh, M. H., Thebault, S., et al. (2007). Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J. Virol. 81, 1543–1553. doi: 10.1128/JVI.00480-06
Lezin, A., Gillet, N., Olindo, S., Signate, A., Grandvaux, N., Verlaeten, O., et al. (2007). Histone deacetylase-mediated transcriptional activation reduces proviral loads in HTLV-1-associated myelopathy/tropical spastic paraparesis patients. Blood 110, 3722–3728. doi: 10.1182/blood-2007-04-085076
Ma, G., Yasunaga, J., Akari, H., and Matsuoka, M. (2015). TCF1 and LEF1 act as T-cell intrinsic HTLV-1 antagonists by targeting Tax. Proc. Natl. Acad. Sci. U.S.A. 112, 2216–2221. doi: 10.1073/pnas.1419198112
Ma, G., Yasunaga, J., and Matsuoka, M. (2016). Multifaceted functions and roles of HBZ in HTLV-1 pathogenesis. Retrovirology 13:16. doi: 10.1186/s12977-016-0249-x
MacNamara, A., Rowan, A., Hilburn, S., Kadolsky, U., Fujiwara, H., Suemori, K., et al. (2010). HLA class I binding of HBZ determines outcome in HTLV-1 infection. PLoS Pathog. 6:e1001117. doi: 10.1371/journal.ppat.1001117
Maertens, G. N. (2016). B’-protein phosphatase 2A is a functional binding partner of delta-retroviral integrase. Nucleic Acids Res. 44, 364–376. doi: 10.1093/nar/gkv1347
Manel, N., Kim, F. J., Kinet, S., Taylor, N., Sitbon, M., and Battini, J. L. (2003). The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 115, 449–459. doi: 10.1016/S0092-8674(03)00881-X
Matsuzaki, T., Nakagawa, M., Nagai, M., Usuku, K., Higuchi, I., Arimura, K., et al. (2001). HTLV-I proviral load correlates with progression of motor disability in HAM/TSP: analysis of 239 HAM/TSP patients including 64 patients followed up for 10 years. J. Neurovirol. 7, 228–234. doi: 10.1080/13550280152403272
Melamed, A., Laydon, D. J., Gillet, N. A., Tanaka, Y., Taylor, G. P., and Bangham, C. R. (2013). Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog. 9:e1003271. doi: 10.1371/journal.ppat.1003271
Merkenschlager, M., and Nora, E. P. (2016). CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17, 17–43. doi: 10.1146/annurev-genom-083115-022339
Miyazato, P., Matsuo, M., Katsuya, H., and Satou, Y. (2016). Transcriptional and epigenetic regulatory mechanisms affecting HTLV-1 provirus. Viruses 8:E171. doi: 10.3390/v8060171
Nagai, M., Usuku, K., Matsumoto, W., Kodama, D., Takenouchi, N., Moritoyo, T., et al. (1998). Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J. Neurovirol. 4, 586–593. doi: 10.3109/13550289809114225
Nakano, K., and Watanabe, T. (2012). HTLV-1 Rex: the courier of viral messages making use of the host vehicle. Front. Microbiol. 3:330. doi: 10.3389/fmicb.2012.00330
Nyborg, J. K., Egan, D., and Sharma, N. (2010). The HTLV-1 Tax protein: revealing mechanisms of transcriptional activation through histone acetylation and nucleosome disassembly. Biochim. Biophys. Acta 1799, 266–274. doi: 10.1016/j.bbagrm.2009.09.002
Olindo, S., Belrose, G., Gillet, N., Rodriguez, S., Boxus, M., Verlaeten, O., et al. (2011). Safety of long-term treatment of HAM/TSP patients with valproic acid. Blood 118, 6306–6309. doi: 10.1182/blood-2011-04-349910
Pais-Correia, A. M., Sachse, M., Guadagnini, S., Robbiati, V., Lasserre, R., Gessain, A., et al. (2010). Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat. Med. 16, 83–89. doi: 10.1038/nm.2065
Parker, C. E., Daenke, S., Nightingale, S., and Bangham, C. R. (1992). Activated, HTLV-1-specific cytotoxic T-lymphocytes are found in healthy seropositives as well as in patients with tropical spastic paraparesis. Virology 188, 628–636. doi: 10.1016/0042-6822(92)90517-S
Pise-Masison, C. A., Castro-Amarante, M. F. D., Enose-Akahata, Y., Buchmann, R. C., Fenizia, C., Washington, P. R., et al. (2014). Co-dependent of HTLV-1 p12 and p8 function in virus persistence. PLoS Pathog. 10:e1004454. doi: 10.1371/journal.ppat.1004454
Rende, F., Cavallari, I., Corradin, A., Silic-Benussi, M., Toulza, F., Toffolo, G. M., et al. (2011). Kinetics and intracellular compartmentalization of HTLV-1 gene expression: nuclear retention of HBZ mRNAs. Blood 117, 4855–4859. doi: 10.1182/blood-2010-11-316463
Robek, M. D., Wong, F. H., and Ratner, L. (1998). Human T-cell leukemia virus type 1 pX-I and pX-II open reading frames are dispensable for the immortalization of primary lymphocytes. J. Virol. 72, 4458–4462.
Rosewick, N., Durkin, K., Artesi, M., Marcais, A., Hahaut, V., Griebel, P., et al. (2017). Cis-perturbation of cancer drivers by the HTLV-1/BLV proviruses is an early determinant of leukemogenesis. Nat. Commun. 8:15264. doi: 10.1038/ncomms15264
Satou, Y., Miyazato, P., Ishihara, K., Yaguchi, H., Melamed, A., Miura, M., et al. (2016). The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome. Proc. Natl. Acad. Sci. U.S.A. 113, 3054–3059. doi: 10.1073/pnas.1423199113
Sharma, N., and Nyborg, J. K. (2008). The coactivators CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter. Proc. Natl. Acad. Sci. U.S.A. 105, 7959–7963. doi: 10.1073/pnas.0800534105
Shiohama, Y., Naito, T., Matsuzaki, T., Tanaka, R., Tomoyose, T., Takashima, H., et al. (2016). Absolute quantification of HTLV-1 basic leucine zipper factor (HBZ) protein and its plasma antibody in HTLV-1 infected individuals with different clinical status. Retrovirology 13:29. doi: 10.1186/s12977-016-0263-z
Silic-Benussi, M., Cavallari, I., Vajente, N., Vidali, S., Chieco-Bianchi, L., Di Lisa, F., et al. (2010). Redox regulation of T-cell turnover by the p13 protein of human T-cell leukemia virus type 1: distinct effects in primary versus transformed cells. Blood 116, 54–62. doi: 10.1182/blood-2009-07-235861
Silverman, L. R., Phipps, A. J., Montgomery, A., Ratner, L., and Lairmore, M. D. (2004). Human T-cell lymphotropic virus type 1 open reading frame II-encoded p30II is required for in vivo replication: evidence of in vivo reversion. J. Virol. 78, 3837–3845. doi: 10.1128/JVI.78.8.3837-3845.2004
Spina, C. A., Anderson, J., Archin, N. M., Bosque, A., Chan, J., Famiglietti, M., et al. (2013). An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 9:e1003834. doi: 10.1371/journal.ppat.1003834
Suzuki, T., Fujisawa, J. I., Toita, M., and Yoshida, M. (1993). The trans-activator tax of human T-cell leukemia virus type 1 (HTLV-1) interacts with cAMP-responsive element (CRE) binding and CRE modulator proteins that bind to the 21-base-pair enhancer of HTLV-1. Proc. Natl. Acad. Sci. U.S.A. 90, 610–614. doi: 10.1073/pnas.90.2.610
Taniguchi, Y., Nosaka, K., Yasunaga, J., Maeda, M., Mueller, N., Okayama, A., et al. (2005). Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 2:64. doi: 10.1186/1742-4690-2-64
Tripathy, M. K., Mcmanamy, M. E. M., Burch, B. D., Archin, N. M., and Margolis, D. M. (2015). H3K27 demethylation at the proviral promoter sensitizes latent HIV to the effects of vorinostat in ex vivo cultures of resting CD4+ T cells. J. Virol. 89, 8392–8405. doi: 10.1128/JVI.00572-15
Tsai, A. G., Cabrales, P., Hangai-Hoger, N., and Intaglietta, M. (2004). Oxygen distribution and respiration by the microcirculation. Antioxid. Redox Signal. 6, 1011–1018. doi: 10.1089/ars.2004.6.1011
Valeri, V. W., Hryniewicz, A., Andresen, V., Jones, K., Fenizia, C., Bialuk, I., et al. (2010). Requirement of the human T-cell leukemia virus p12 and p30 products for infectivity of human dendritic cells and macaques but not rabbits. Blood 116, 3809–3817. doi: 10.1182/blood-2010-05-284141
Yasunaga, J.-I., Furuta, R., Miura, M., Sugata, K., Saito, A., Hirofumi, A., et al. (2016). Hematopoietic stem cell infected with HTLV-1 functions as a viral reservoir in vivo. Blood 128, 1343–1343.
Keywords: HTLV-1, latency, virus, metabolism, hypoxia, glucose, epigenetic regulation, transcription
Citation: Kulkarni A and Bangham CRM (2018) HTLV-1: Regulating the Balance Between Proviral Latency and Reactivation. Front. Microbiol. 9:449. doi: 10.3389/fmicb.2018.00449
Received: 22 December 2017; Accepted: 27 February 2018;
Published: 19 March 2018.
Edited by:
Umberto Bertazzoni, University of Verona, ItalyReviewed by:
Luc Willems, Fonds National de la Recherche Scientifique, BelgiumDonna M. D’Agostino, Università degli Studi di Padova, Italy
Copyright © 2018 Kulkarni and Bangham. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles R. M. Bangham, c.bangham@imperial.ac.uk
 Anurag Kulkarni
Anurag Kulkarni Charles R. M. Bangham
Charles R. M. Bangham