- 1Department of Microbiology and Immunology, Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 2Scientific Research Center of Wenzhou Medical University, Wenzhou, China
Swarming is a surface-associated motile behavior that plays an important role in the rapid spread, colonization, and subsequent establishment of bacterial communities. In Pseudomonas aeruginosa, swarming is dependent upon a functional flagella and aided by the production of biosurfactants. AmrZ, a conserved transcription factor across pseudomonads, has been shown to be a global regulator of multiple genes important for virulence and ecological fitness. In this study, we expand this concept of global control to swarming motility by showing that deletion of amrZ results in a severe defect in swarming, while multicopy expression of this gene stimulates swarming of P. aeruginosa. Mechanistic studies showed that the swarming defect of an amrZ mutant does not involve changes of biosurfactant production but is associated with flagellar malfunction. The ∆amrZ mutant exhibits increased levels of the second messenger cyclic di-GMP (c-di-GMP) compared to the wild-type strain, under swarming conditions. We found that the diguanylate cyclase GcbA was the main contributor to the increased accumulation of c-di-GMP observed in the ∆amrZ mutant and was a strong inhibitor of flagellar-dependent motility. Our results revealed that the GcbA-dependent inhibition of motility required the presence of two c-di-GMP receptors containing a PilZ domain: FlgZ and PA14_56180. Furthermore, the ∆amrZ mutant exhibits enhanced production of Pel polysaccharide. Epistasis analysis revealed that GcbA and the Pel polysaccharide act independently to limit swarming in ΔamrZ. Our results support a role for AmrZ in controlling swarming motility, yet another social behavior besides biofilm formation that is crucial for the ability of P. aeruginosa to colonize a variety of surfaces. The central role of AmrZ in controlling these behaviors makes it a good target for the development of treatments directed to combat P. aeruginosa infections.
Introduction
Swarming is a surface-associated mode of motility that involves rapid and coordinated movement of a bacterial population across viscous semisolid surfaces (Kearns, 2010; Partridge and Harshey, 2013). In the opportunistic human pathogen Pseudomonas aeruginosa (Silby et al., 2011), this type of movement requires the presence of a functional flagella to mediate actual movement and is aided by the production of biosurfactants such as rhamnolipids to overcome the surface tension between cells and the surrounding environment (Kohler et al., 2000; Kearns, 2010). Since human mucosal surfaces, such as the epithelial surfaces of the lung represents a viscous environment analogous to the conditions that promote swarming in vitro, swarming motility is considered clinically relevant (Yeung et al., 2012). Moreover, there is evidence that swarming does not simply enable the bacterium to move but is also a complex lifestyle adaptation in response to various environmental cues, resulting in substantial changes in metabolism (Yeung et al., 2009; Ghorbal et al., 2019), increased virulence gene expression and antibiotic resistance (Overhage et al., 2008; Tremblay and Deziel, 2010). Thus, investigation of the key molecules and mechanisms regulating swarming motility is important for the development of treatment against this bacterium.
Cyclic di-GMP (c-di-GMP) is a nearly ubiquitous bacterial second messenger that regulates diverse cellular processes and is of key importance for modulating transitions between motile and sessile lifestyles important for acute and chronic infections, respectively (Hengge, 2009; Romling et al., 2013). Like other bacteria, P. aeruginosa also uses c-di-GMP to create an inverse regulation of biofilm formation and swarming motility (Valentini and Filloux, 2016; Baker et al., 2019). According to a well-established model, increased levels of c-di-GMP have a negative effect on swarming and are correlated with a sessile lifestyle, while at low levels the bacteria can move and swarm away in search of better conditions (Ha and O’Toole, 2015; Yan et al., 2017). c-di-GMP is synthesized by diguanylate cyclases (DGCs) and is degraded by phosphodiesterases (PDEs). The P. aeruginosa genome encodes over 40 such enzymes that contribute to the steady-state levels of intracellular c-di-GMP (Ha and O’Toole, 2015). To exert its control, c-di-GMP binds to different classes of effector proteins or RNAs (Hengge, 2009). Currently, proteins with a PilZ domain represent the largest family of c-di-GMP effectors (Orr and Lee, 2016), and the highly conserved RXXXR and (D/N)XSXXG motifs in the PilZ domain are essential for c-di-GMP binding (Amikam and Galperin, 2006; Cheang et al., 2019). In the c-di-GMP-bound state, PilZ domain proteins regulate diverse cellular processes such as virulence, biofilm formation and flagellum-dependent motility (Baker et al., 2016; Xu et al., 2016a,b).
AmrZ, a transcription factor that belongs to the ribbon-helix-helix (RHH) family of DNA-binding proteins (Pryor et al., 2012), was originally described as AlgZ for its ability to activate alginate production in P. aeruginosa (Baynham and Wozniak, 1996). It was later changed to AmrZ (alginate and motility regulator) because of its positive role in the regulation of Type IV Pili biogenesis and twitching motility (a pili dependent motility) (Baynham et al., 2006). Its implication in regulation of motility was then expanded by the finding that AmrZ acts as a negative regulator of flagellum biosynthesis in mucoid, nonmotile P. aeruginosa isolates from CF patients (Tart et al., 2006). In this case, AmrZ is expressed at high levels and directly represses transcription of the flagellar master regulator FleQ and thus flagellum biosynthesis (Tart et al., 2006). However, in nonmucoid strains, the synthesis of flagellin is similar between wild-type and the amrZ deletion mutant (Baynham et al., 2006), and the role of AmrZ in flagellum-driven motility is still unclear. Additionally, AmrZ inhibits the production of the Psl polysaccharide by directly repressing transcription of the psl operon, which is involved in P. aeruginosa biofilm development (Jones et al., 2013, 2014). Recent studies have shown that AmrZ functions as a global regulator (Jones et al., 2014; Martinez-Granero et al., 2014; Allsopp et al., 2017) and one of the AmrZ regulated targets is c-di-GMP signaling. It was shown that the ΔamrZ mutant displays elevated levels of c-di-GMP and forms hyper biofilms compared with the wild-type strain PAO1 (Jones et al., 2014). Considering that c-di-GMP levels are higher in ΔamrZ and that elevated levels of c-di-GMP inhibit motility (McCarter and Gomelsky, 2015), we hypothesized motility might be impaired in ΔamrZ cells.
Here, we investigated the role of AmrZ in swarming and the underlying molecular mechanism of this association. We find that AmrZ positively controls swarming motility of P. aeruginosa and the swarming deficiency of a ∆amrZ mutant is associated with flagellar malfunction but not insufficient biosurfactant production. We demonstrate that the AmrZ-mediated regulation of swarming involves a c-di-GMP signaling module consisting of the DGC GcbA and the c-di-GMP receptors FlgZ and PA14_56180. We also uncovered negative regulation of Pel polysaccharide production by AmrZ. This regulation does not involve changes in pel expression and modulates the ability of P. aeruginosa to swarm.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
The strains and plasmids used in this study are listed in Supplementary Table S1. P. aeruginosa strain UCBPP-PA14 (abbreviated P. aeruginosa PA14) was used as the parental strain, unless otherwise stated. Escherichia coli DH5α was used as the host for DNA cloning. P. aeruginosa and E. coli strains were routinely cultured in Lysogeny Broth (LB) medium (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter, pH 7.0) or on LB Agar (LB medium containing 1.5% [w/v] agar) at 37°C unless otherwise noted. For expression plasmids with the PBAD promoter, arabinose was added to cultures at a 0.2% final concentration. Where necessary, 50 μg/ml gentamicin, 150 μg/ml carbenicillin, and 50 μg/ml tetracycline were used for P. aeruginosa and 100 μg/ml ampicillin, 50 μg/ml kanamycin, 10 μg/ml gentamicin, and 10 μg/ml tetracycline were used for E. coli.
Construction of Strains and Plasmids
In-frame deletion mutants were constructed by allelic exchange using the sucrose counter-selection system as previously described with the P. aeruginosa suicide vector pEX18Tc (Hoang et al., 1998). Mutant strains were confirmed by PCR analysis of genomic DNA. In addition, single-copy chromosomal complementation of the amrZ mutation was accomplished by introducing amrZ under the control of its native promoter into pUC18T-mini-Tn7T-Gm (Choi and Schweizer, 2006). Overexpression was accomplished by placing the respective genes under the control of the constitutive lac promoter in pUCP20 (West et al., 1994). For complementation of the flgZ or PA14_56180 mutant in the GcbA-overexpressing background with the respective C-terminally FLAG-tagged WT and mutated proteins, primers were designed to contain the open reading frame and ribosome-binding site with the indicated oligonucleotides (Supplementary Table S2) and expression of the respective genes was under the control of the arabinose-inducible PBAD promoter in the pUC18T-mini-Tn7T-Gm-BAD integration vector. pUC18T-mini-Tn7T-Gm-BAD was created by inserting a NsiI-SacI digested araC-PBAD cassette from pBAD18 (Guzman et al., 1995) into pUC18T-mini-Tn7T-Gm cleaved at its NsiI and SacI sites in the MCS. The identity of vector inserts was verified by PCR and sequencing. Plasmids were introduced into P. aeruginosa by electroporation (Choi et al., 2006). The primers used for strain construction are listed in Supplementary Table S2.
Motility Assays
Swarming motility assays were performed as previously described on 0.5% (w/v) agar M8 plates (Ha et al., 2014) supplemented with 0.2% (w/v) glucose, 0.5% (w/v) casamino acids, and 1 mM MgSO4 (Kohler et al., 2000; Kuchma et al., 2007). After solidification, plates were briefly dried at room temperature and spot inoculated with 2.5 μl aliquots taken directly from overnight LB cultures. Swarming plates were incubated face up in stacks of no more than two at 37°C for 16–18 h. To quantify the degree of swarming, an image of the swarming plate was captured with a digital camera and percent coverage of the plate was measured by comparing swarming pixels with total plate pixels using Adobe Photoshop CS6. Swim plates were identical to swarm plates except that it was solidified with 0.3% (w/v) agar (Caiazza et al., 2005). Swimming assays were carried out as reported previously and incubated at 30°C for 18–20 h (Murray et al., 2010; Li et al., 2017). Experiments were repeated in triplicate and the data are presented as averages over three replicate plates.
Quantitative Real-Time Reverse-Transcription PCR Analysis
Quantitative real-time reverse-transcription PCR (qRT-PCR) was used to determine the gene expression levels of wild-type and indicated mutant strains. Planktonic cells were grown in liquid swarming medium to the exponential phase (OD600 = 0.9–1.0) under shaking conditions (Overhage et al., 2008). Cells growing on swarming plate were harvested from the tip of migrating tendrils or from the center of swarming colonies and were collected directly into RNAlater reagent (Qiagen, Germany) (Overhage et al., 2008; Tremblay and Deziel, 2010). Biofilms were grown directly on 6-well polystyrene microplates (Costar #3506) in liquid swarming medium and incubated as a static culture for 6 and 24 h at 37°C (Friedman and Kolter, 2004; Petrova et al., 2014; Price et al., 2016). At each time point, unattached bacteria were removed by aspiration and the remaining biofilms were washed once with PBS and disrupted with a cell scraper. RNA was extracted using the RNAprep pure Kit (TIANGEN Biotech, Beijing, China) according to the manufacturer’s instructions. To remove residual DNA, RNA was further treated with DNase I, repurified with an RNAclean Kit (TIANGEN), and was confirmed to be free of DNA by PCR. cDNA was synthesized with a PrimeScript RT Reagent Kit (Perfect Real Time) (TaKaRa, Liaoning, China) using random hexamer primers. qRT-PCR was performed on the Bio-Rad CFX96 Touch real-time PCR detection system using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) mix (Takara). The mRNA levels were normalized by the geometric mean of PA14_26910 (PA2875) and PA14_20860 (PA3340) transcript levels (Costaglioli et al., 2014), and relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Rhamnolipid Production and Drop Collapse Assays
Rhamnolipid biosynthesis was analyzed via the cetyltrimethylammonium bromide (CTAB)-methylene blue plate method as previously described (Siegmund and Wagner, 1991; Caiazza et al., 2005). Briefly, 2.5 μl of overnight LB cultures were spot inoculated onto plates consisting of the same base medium as for the swarming assays but supplemented with 0.02% (w/v) cetyltrimethylammonium bromide (CTAB), 0.0005% (w/v) methylene blue, and solidified with 1.5% (w/v) agar. Plates were incubated face up at 37°C for 24 h, and then incubated for an additional 24 h at room temperature. Rhamnolipid production was determined by measuring the diameter of the dark blue halo surrounding a colony. To detect the production of the rhamnolipid precursor HAA, drop collapse assays were performed on 0.22-μm membrane filtered supernatants prepared from LB overnight cultures at 30°C (Murray and Kazmierczak, 2008). Filtered supernatant was serially diluted (1:1) with sterile water in a 96-well plate, and 30 μl of each dilution was spotted onto the circles on the underside of the lid of a Corning 96-well plate, allowing surfactant activity to be measured by the spread of the droplet as described (Caiazza et al., 2005; Murray and Kazmierczak, 2008). Each assay was repeated at least three times.
Transmission Electron Microscopy
Transmission electron microscopy (TEM) was used to visualize cell morphology and the presence of flagella as described previously (Kohler et al., 2000; Rashid and Kornberg, 2000). Briefly, bacteria at the advancing swarm edge were gently scraped and suspended in 2.5% (v/v) glutaraldehyde in 0.1 M PBS (pH 7.2) and the fixation process started at the same time. Fifteen minutes later, carbon-coated copper grids were placed on top of a drop of the cell suspension for 30 s to allow the adhesion of bacterial cells. Grids were then stained for 2 s with 2% uranyl acetate (w/v) and washed twice for 10 s in a drop of distilled water. The grid was air dried and examined on a Hitachi H-7500 transmission electron microscope at calibrated magnifications. Approximately 200 cells were counted to determine the percentage of flagellated cells for each strain.
Reversal Rate Measurements
Reversal rate measures the frequency at which a motile cell changes its direction (Toutain et al., 2005; Caiazza et al., 2007; Petrova et al., 2014). Bacteria expressing green fluorescent protein (GFP) were cultured overnight, sub-cultured and grown to the exponential phase (OD600 of 1.0) before observation. The GFP-expressing bacteria were generated by introduction of a multicopy plasmid (pUCP-zsGreen1, GFP+) that constitutively expresses ZsGreen1 GFP under the control of the lac promoter (Li et al., 2017). Cultures were then diluted 1:100 into M63 medium (Kuchma et al., 2007) supplemented with 0.2% (w/v) glucose and containing 3% (w/v, low-viscosity/swimming conditions) or 15% (w/v) Ficoll (high-viscosity/swarming conditions) (Toutain et al., 2005; Caiazza et al., 2007). Cells were monitored via fluorescence microscope (Eclipse Ni-U, Nikon) equipped with a 100×/1.45 oil objective lens and a Nikon DS-Fi1C camera. Real-time videos were captured using the NIS-Elements F Ver4.00.00 and Camtasia Studio V7.5 software package. The videos were subsequently analyzed to monitor individual cells for the number of times they reversed motility direction while within the field of view. Approximately 50 cells were measured for each strain and reversal rates are expressed as no. of reversals per cell per minute.
Cyclic di-GMP Quantification
Intracellular levels of c-di-GMP were determined by LC-MS/MS as previously described with minor modifications (Merritt et al., 2010; Spangler et al., 2010; Chen et al., 2014). Swarm cells were washed off the surface of swarming plates with PBS by gently pipetting while slightly inclining the petri dish. Six swarming plates were used for each strain and for the swarming deficient amrZ mutant, each plate was inoculated with three colonies and 10 plates were used. Cell numbers were then normalized to an OD600 of 2.0, and 5 ml of this suspension was pelleted by centrifugation at 4°C for 10 min at 5,000 rpm. Pellets were resuspended in a small amount of supernatant, transferred to a pre-weighed 1.5 ml microcentrifuge tube, and then pelleted again. After removing the supernatant, cell pellets were vigorously resuspended in 250 μl of extraction buffer (acetonitrile/methanol/water [40/40/20, v/v/v] plus 0.1 M formic acid) and incubated at −20°C for 30 min. The cell debris was then centrifuged at 13,000 rpm for 5 min at 4°C, and 200 μl of supernatants containing the nucleotide extract were recovered into new tubes and neutralized with 4 μl of 15% NH4HCO3 (w/v) per 100 μl of sample. The resultant extracts and the tubes with cell debris were dried using a vacuum concentrator. The pellet weights were measured to get sample dry weight, and the dried extracts were resuspended in 200 μl of HPLC grade water and analyzed by LC-MS/MS to quantify the amount of c-di-GMP. Quantifications were performed in triplicate and the c-di-GMP levels were normalized to the dry weight of the cell pellet from which c-di-GMP was extracted and presented as pmol of c-di-GMP/mg of dry weight (Kuchma et al., 2012; Chen et al., 2014).
Western Blot Analysis
Strains were cultured on swarming plates supplemented with 0.2% arabinose and appropriate antibiotics (Kuchma et al., 2015). Cells were harvested by gentle scraping with plastic coverslips and resuspended in PBS. Samples were normalized to equivalent number of bacteria cells by OD600 (Li et al., 2013). For Western blotting, whole-cell lysates were resolved by SDS-PAGE using 15% polyacrylamide gels. Proteins transferred to a polyvinylidene difluoride (PVDF) membrane were probed with an anti-FLAG antibody (Sigma, Shanghai, China) to detect FLAG-tagged FlgZ and PA14_56180 and mutant variants of these proteins. Signals were developed with a Western Lightning Plus-ECL kit (PerkinElmer, MA, USA).
Congo Red-Binding Assays
To examine exopolysaccharide production, Congo red (CR)-binding assays were performed on tryptone (10 g/L) agar (1%) plates supplemented with CR (40 μg/ml) and Coomassie brilliant blue R (20 μg/ml) (Kong et al., 2013). Bacteria from LB-grown overnight cultures were diluted to an OD600 of 0.025 in PBS and a 2 μl volume was used for inoculation. Plates were incubated at 37°C for 24 h, followed by 4 days at room temperature before inspection (Kuchma et al., 2007; Colvin et al., 2012). To quantify the CR-binding levels (Andrews and Maurelli, 1992; Cangelosi et al., 1999), colonies were scraped from CR plates and suspended with vigorous vortexing in 600 μl of deionized water. After recording the OD660 of the suspension, cells were pelleted, resuspended in 200 μl of acetone, vortexed and the CR dye was extracted at room temperature for 2 h. Cells were then centrifuged, and the OD488 of the supernatant containing the extracted CR was measured. The relative Congo red-binding (RCRB) value was calculated by dividing the OD488 of the acetone extracts by the OD660 of the original cell suspension.
Data Presentation and Statistical Analysis
The data are presented as the mean values ± standard deviations (mean ± SD). All significant differences were evaluated on SPSS program by using the Student’s t-test or one-way analysis of variance (ANOVA) followed by the Tukey’s test for multiple comparisons when applicable: *p < 0.05; **p < 0.01.
Results
AmrZ Is an Important Contributor to Swarming Motility in P. aeruginosa
To evaluate the potential role of AmrZ in the swarming motility of P. aeruginosa, we initially constructed an in-frame deletion of amrZ in strain PA14 and examined its swarming phenotype. As shown in Figure 1A, deletion of the amrZ gene resulted in a severe defect in swarming motility compared to the parental wild-type (WT) strain. The amrZ mutant grows at a rate indistinguishable from that of the WT under the same medium conditions (data not shown), suggesting the swarming defect observed was not due to decreased growth rate.
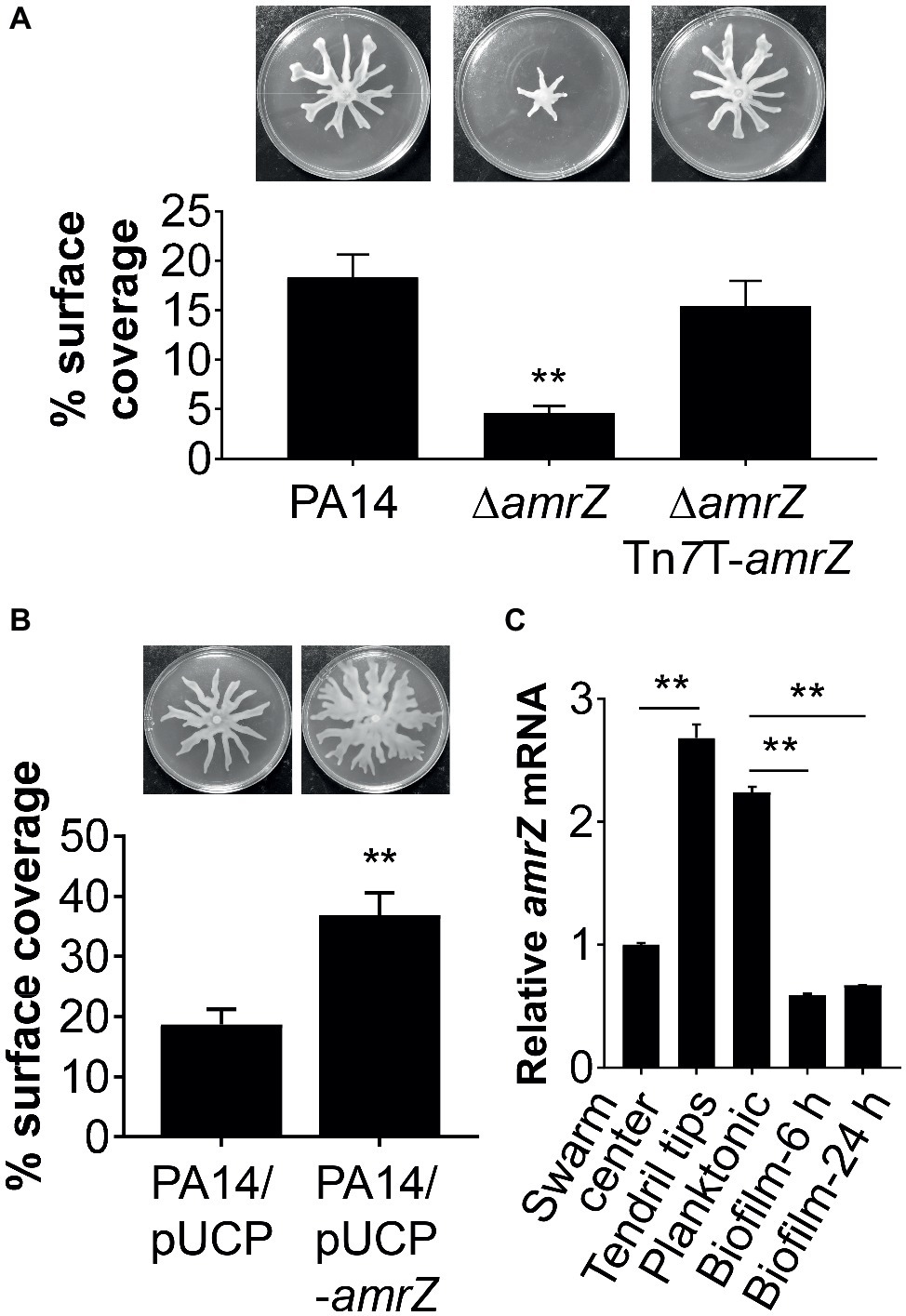
Figure 1. AmrZ plays a role in the regulation of swarming motility in P. aeruginosa PA14. (A) Swarming motility and average percentage of the surface coverage of WT PA14, the ΔamrZ mutant, and the single-copy complemented strain of the ΔamrZ mutant carrying an amrZ gene inserted into chromosome by mini-Tn7T. Error bars represent the standard deviations. **p < 0.01 compared with WT or complemented strains by Student’s t-test. (B) Swarm analysis and quantification of percent coverage of swarm plates for the WT strain carrying vector control (PA14/pUCP) or a multicopy amrZ-containing plasmid (PA14/pUCP-amrZ). (C) RT-qPCR analysis of amrZ mRNA levels in WT PA14 from actively swarming tendril tips, cells localized in the center of swarming colonies, free-swimming planktonic cells, and cells grown as biofilm at 6 h (initial attachment) and 24 h (developed biofilm). The relative gene expression was reported relative to the levels in swarm center cells. Significance was determined by Students’ t-test (**p < 0.01).
To confirm that the swarming defect of the ∆amrZ mutant was indeed due to the loss of amrZ, we generated a complementation construct (pTn7T-amrZ) to express the amrZ gene under its own promoter at the chromosomal attTn7 site (Choi and Schweizer, 2006). As shown in Figure 1A, complementation of the amrZ mutant restored the swarming phenotype to WT levels, demonstrating the observed swarming defect is due to the amrZ deletion. Moreover, multicopy expression of amrZ (pUCP-amrZ) in WT PA14 resulted in a hyperswarming phenotype compared to that of the vector control (Figure 1B), reinforcing the finding that AmrZ positively regulates swarming motility of P. aeruginosa. In addition, deletion of amrZ in the type strain PAO1 also resulted in a strong impairment in swarming motility, and the swarming defective phenotype could be restored to WT levels by native expression of the chromosomally integrated amrZ-PAO1 gene (Supplementary Figure S1), suggesting the influence of AmrZ on swarming motility is not strain-specific.
Recent findings suggest that distinct bacterial subpopulations are present within a swarming colony (Tremblay and Deziel, 2010). While cells at the tendril tips are fast moving and rapidly spread on uncolonized surfaces, swarm center populations live in a biofilm-like state allowing a permanent settlement of the colonized areas (Tremblay and Deziel, 2010). To further understand the role of AmrZ in lifestyle adaptation, especially its association with swarming behavior, we analyzed the expression pattern of amrZ in swarming colonies of WT PA14 located at the tendril tips, swarming center as well as cells grown planktonically or as biofilms at both 6 h (initial attachment) and 24 h (developed biofilm). It was found that amrZ expression in actively swarming tendril tips was upregulated 2.68-fold compared to that in swarming center cells (Figure 1C), and the expression in swarming tips was comparable to that of cells grown planktonically (Figure 1C). The expression of amrZ in biofilm cells was downregulated 3.8- and 3.35-fold at 6 and 24 h, respectively, relative to their planktonic counterparts and this decrease was more striking when compared to swarming tendril tip cells (Figure 1C), indicating the expression of amrZ in motile cells was higher than that in sessile cells. As loss of amrZ leads to a hyper-biofilm phenotype in P. aeruginosa (Jones et al., 2013), these results support the notion that AmrZ may serve as a molecular switch that regulates the transition between motile and sessile lifestyles. Collectively, these data suggest that AmrZ is an important contributor to swarming motility, and its expression level is closely correlated with the motile and sessile behaviors of P. aeruginosa.
AmrZ-Mediated Swarming Regulation Does Not Implicate Biosurfactant Production but Is Associated With Flagellar Function
Swarming behavior in P. aeruginosa is dependent upon a functional flagellum and the production of biosurfactants (mostly rhamnolipids) (Kohler et al., 2000; Jean-Pierre et al., 2016). To examine whether the swarming-deficient phenotype of ∆amrZ was related to a lack of biosurfactant synthesis, rhamnolipid production assays were first performed (Caiazza et al., 2005; Yeung et al., 2009). The rhlA mutant was included as a control since this mutant has been shown to be defective in rhamnolipid production (Deziel et al., 2003; Caiazza et al., 2005). We found that the amrZ mutant had a dark blue halo around the colony similar to that for the WT (Figure 2A), while the rhlA mutant had no visible haloes around the colony, indicating that the amrZ mutant was not defective in the production of rhamnolipid. We also measured the production of rhamnolipid precursor 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs) by a drop collapse assay (Caiazza et al., 2005; Murray and Kazmierczak, 2008), and found the ∆amrZ strain showed equivalent drop collapse activity compared to the WT (Figure 2B). Thus, the defect in ΔamrZ mutant swarming is likely not due to a deficiency in HAA and/or rhamnolipid production.
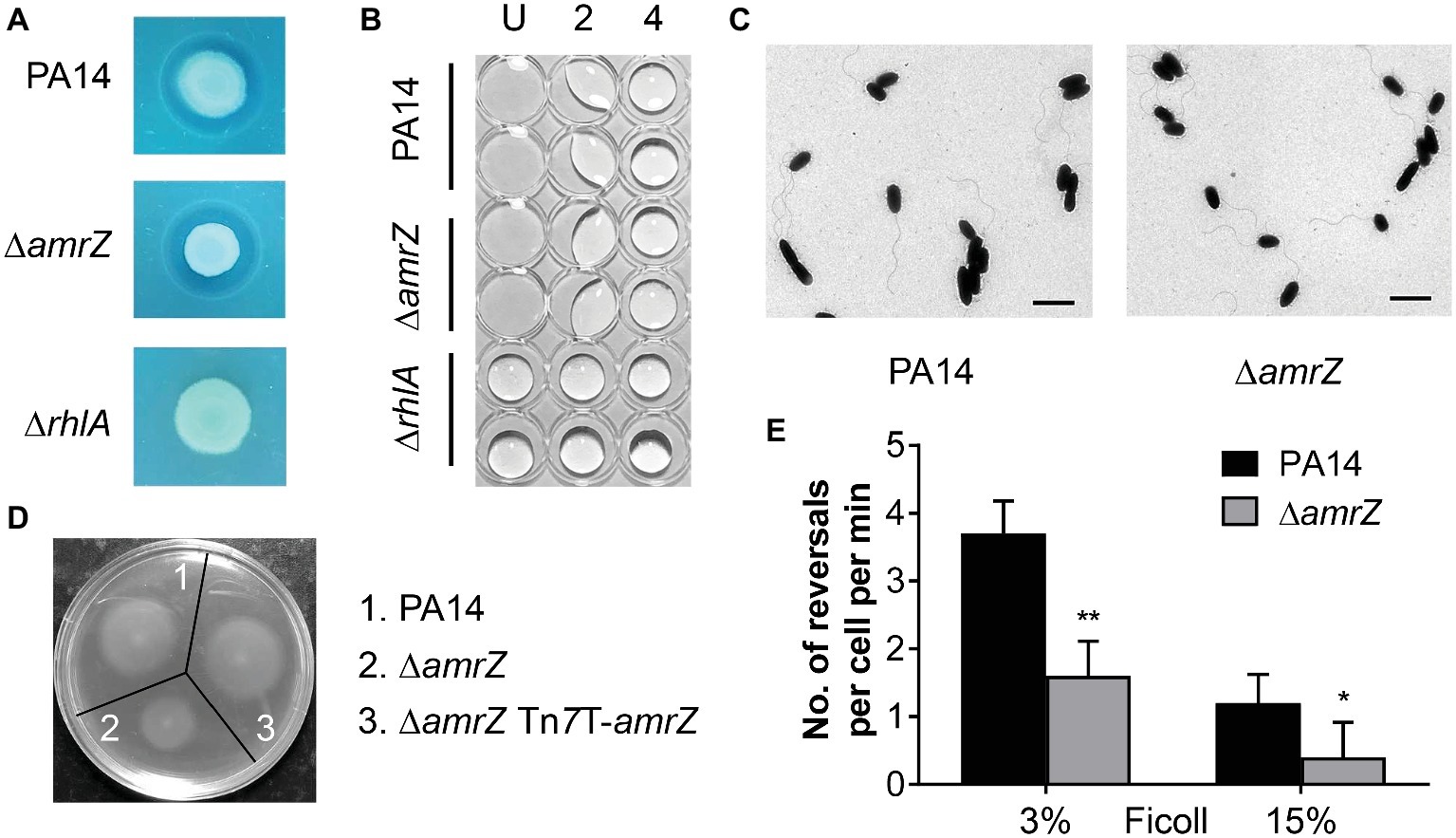
Figure 2. The swarming defect of the amrZ mutant is independent of changes of biosurfactant production or flagellar formation but is associated with flagellar malfunction. (A) Methylene blue-rhamnolipid plate assay. The presence of surfactant is indicated by the formation of a dark ring surrounding a colony and the non-rhamnolipid-producing rhlA mutant served as a negative control. (B) Drop collapse analysis of PA14, ∆amrZ, and ∆rhlA mutants to detect the presence of the rhamnolipid precursor HAA. Samples were diluted in dH2O, spotted on the lid of a 96-well plate, and assayed for bead formation. The dilution factors are shown at the top. U, undiluted. (C) Morphology of PA14 and amrZ mutant cells by TEM of bacteria taken directly from swarm plates. Bar, 2 μm. (D) Swimming motility assay of the indicated strains. (E) Flagellar reversal rates under low (3% Ficoll) and high-viscosity (15% Ficoll) conditions, representing swimming and swarming conditions, respectively, were measured as changes in direction of movement of cells. Rates are expressed as number of reversals per cell per minute. Error bars represent the standard deviations. *p < 0.05; **p < 0.01 compared with WT by Student’s t-test.
Another essential factor that contributes to swarming of P. aeruginosa is flagellum biosynthesis or function. To better understand how AmrZ affects swarming, TEM studies were carried out to determine whether the changes in swarming behavior were due to changes in biosynthesis of the flagellar apparatus. As previously noted for strain PAO1 (Baynham et al., 2006), the amrZ mutant in PA14 possessed well-developed flagella and the percentage of flagellated cells was comparable to that of the WT (88 ± 4% for WT strain and 85 ± 6% for amrZ mutant) (Figure 2C), indicating that there was no defect in flagella formation. We next tested whether flagellar functionality was responsible for the ∆amrZ swarming phenotype by performing swimming motility assays, since swimming motility depends entirely on a functional flagellum (Yeung et al., 2009). As seen in Figure 2D, the ∆amrZ mutant had a severe defect in swimming motility and could be restored by complementation with a functional copy of amrZ, indicating that AmrZ is necessary for flagellar functioning. Furthermore, we examined the effect of AmrZ on flagellar reversal rates in liquid media with viscosities mimicking swimming (3% Ficoll) and swarming (15% Ficoll) conditions (Caiazza et al., 2007). In accord with motility assays on agar plates, inactivation of amrZ resulted in significantly reduced frequency of flagellar reversals under both conditions (Figure 2E), suggesting that AmrZ is involved in flagellar activity. Taken together, these results demonstrate that the swarming repression in the ∆amrZ mutant occurs via flagellar malfunction rather than a default in flagellum biosynthesis or biosurfactant production.
Increased Levels of Cyclic di-GMP Is Partially Responsible for the Swarming Defect of the ∆amrZ Mutant
It has been previously demonstrated that a ∆amrZ mutant exhibited elevated levels of c-di-GMP over that of the parental PAO1 strain (Jones et al., 2014), and it could be expected that c-di-GMP levels will also be higher in a PA14 ∆amrZ mutant strain. However, it was unknown if this would also be the case in swarming cells and whether c-di-GMP was involved in the repression of swarming in ∆amrZ. Therefore, we compared the intracellular levels of c-di-GMP between WT PA14 and the ∆amrZ mutant under swarming conditions. As shown in Figure 3A, c-di-GMP levels were about two times higher in the amrZ mutant than in the WT. This observation is consistent with the previous report by Jones and colleagues (Jones et al., 2014) and coincides with the established notion that high c-di-GMP levels repress swarming motility. To further investigate whether the elevated level of c-di-GMP was responsible for the swarming defect of the ∆amrZ mutant, PA14 and the ∆amrZ mutant strains were transformed with a plasmid encoding the PDE PA2133 (pUCP-2133), which has previously been used to artificially reduce c-di-GMP levels in P. aeruginosa (Moscoso et al., 2011; Frangipani et al., 2014; Li et al., 2017). We found that overexpression of PA2133 led to a significant increase of swarming motility in both WT and ∆amrZ mutant strains (Figures 3B,C). Additionally, we confirmed that WT and ∆amrZ mutant cells overexpressing PA2133 had very low to undetectable levels of c-di-GMP (data not shown). However, as the absence of amrZ still has an effect on swarming even when the PDE is overexpressed, these data suggest that c-di-GMP could be involved in the repression of swarming motility in the ∆amrZ mutant.
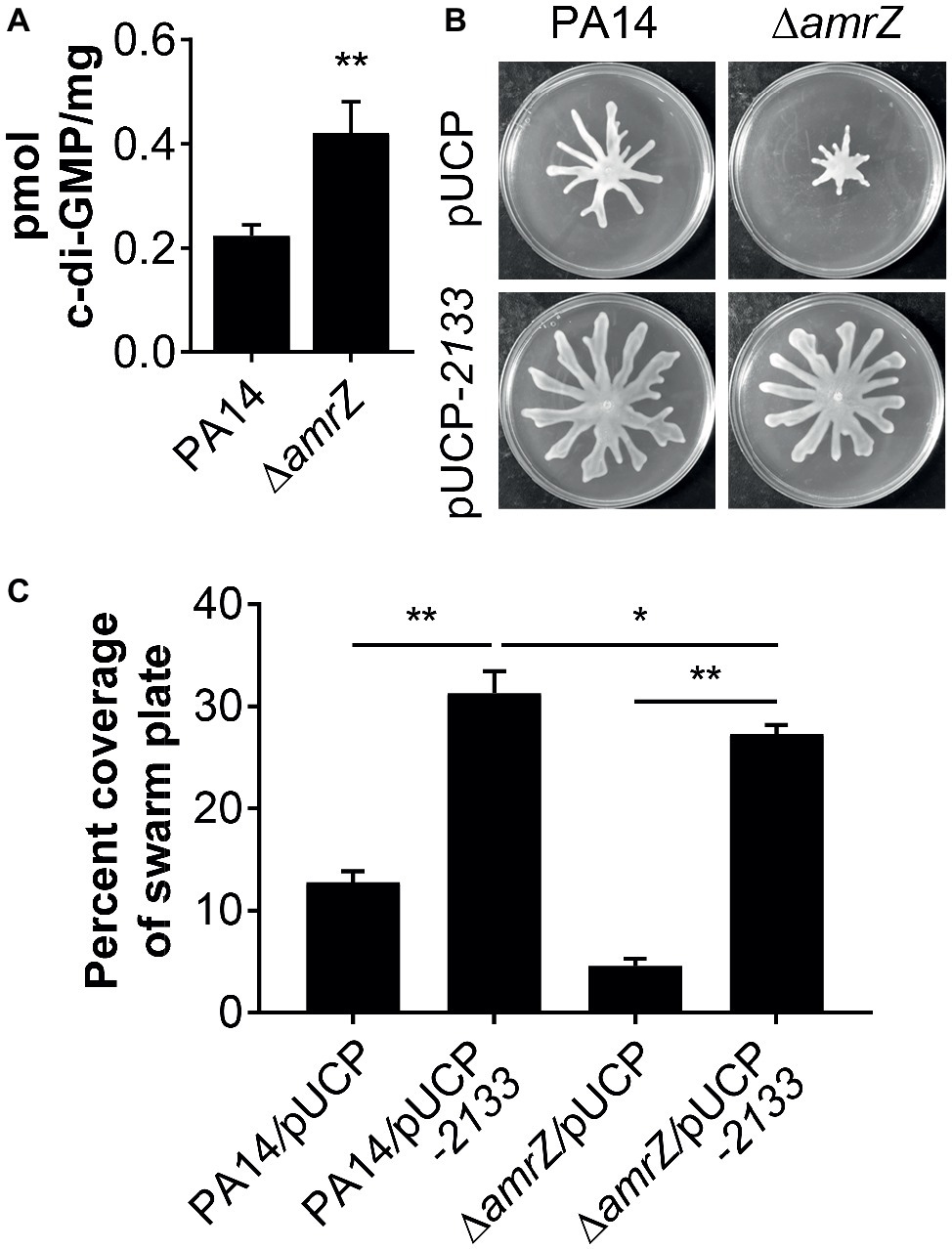
Figure 3. c-di-GMP contributes to the swarming defects of the amrZ mutant. (A) Measurements of c-di-GMP by LC-MS/MS for the indicated strains grown on swarm plates. Data are expressed as pmol of c-di-GMP/mg of dry weight of the cell pellets from which the nucleotides were extracted. (B) Representative swarming images of the WT or amrZ mutant carrying either the empty vector or pUCP-2133 for overproducing the PA2133 phosphodiesterase as indicated. (C) Percent coverage of swarm plates for the strains shown in (B). Statistical analysis is based on three replicates and significance was determined with Student’s t-test (*p < 0.05; **p < 0.01).
The GcbA-Derived Cyclic di-GMP Is Involved in the Swarming Repression Observed in the ΔamrZ Mutant
Our data suggest that there is a c-di-GMP-dependent control of swarming motility in the ∆amrZ mutant. However, as multiple c-di-GMP-metabolizing enzymes exist in P. aeruginosa, we are interested in which DGCs or PDEs could be responsible for the changes in the internal c-di-GMP pool in ∆amrZ and hence swarming motility. In light of the finding that the mRNA expression of the DGC GcbA (also named AdcA) displayed the highest level of upregulation in a transcriptional profiling analysis of a PAO1 amrZ mutant (Jones et al., 2014), we asked whether GcbA plays a role in modulating the c-di-GMP concentration to mediate repression of swarming in ∆amrZ. For the purpose of this study, we validated the expression of gcbA under swarming conditions by qRT-PCR and found the gcbA gene was significantly upregulated (2.89-fold) in ∆amrZ compared to the parental PA14 strain (Figure 4A); however, this upregulation was smaller than that observed in PAO1 ∆amrZ (40.27-fold by RNA-Seq) (Jones et al., 2014). While the RNA-Seq was performed under liquid LBNS (LB medium with no salt) conditions, this result suggests that the viscosity or composition of the environment may cause AmrZ to exert different degrees of downstream effects. We next engineered a ∆amrZ ∆gcbA double mutant and examined its swarming phenotype as well as the levels of c-di-GMP. As a control, a gcbA single mutant was also included. We observed that swarming motility was largely restored in the ∆amrZ ∆gcbA mutant compared to that of the ∆amrZ strain, and the ∆gcbA single mutant also exhibited significantly enhanced swarming relative to the WT (Figures 4B,C). Analysis of the c-di-GMP levels showed that the ∆amrZ ∆gcbA double mutant exhibited significantly reduced c-di-GMP levels compared to that in the ∆amrZ mutant (Figure 4D). However, it is noteworthy that while the ∆amrZ ∆gcbA mutant had a c-di-GMP content at almost the levels of the ΔgcbA single mutant (Figure 4D), swarming motility in the ΔamrZ ΔgcbA mutant was still 2-fold lower than that of the ΔgcbA mutant (Figure 4C); this result implies that AmrZ also affects swarming motility through a gcbA- and (likely) c-di-GMP-independent mechanism. Together these data suggest that GcbA is responsible for the increased production of c-di-GMP in ∆amrZ and the swarming deficiency phenotype of the ∆amrZ mutant is, at least partially, dependent on GcbA-synthesized c-di-GMP.
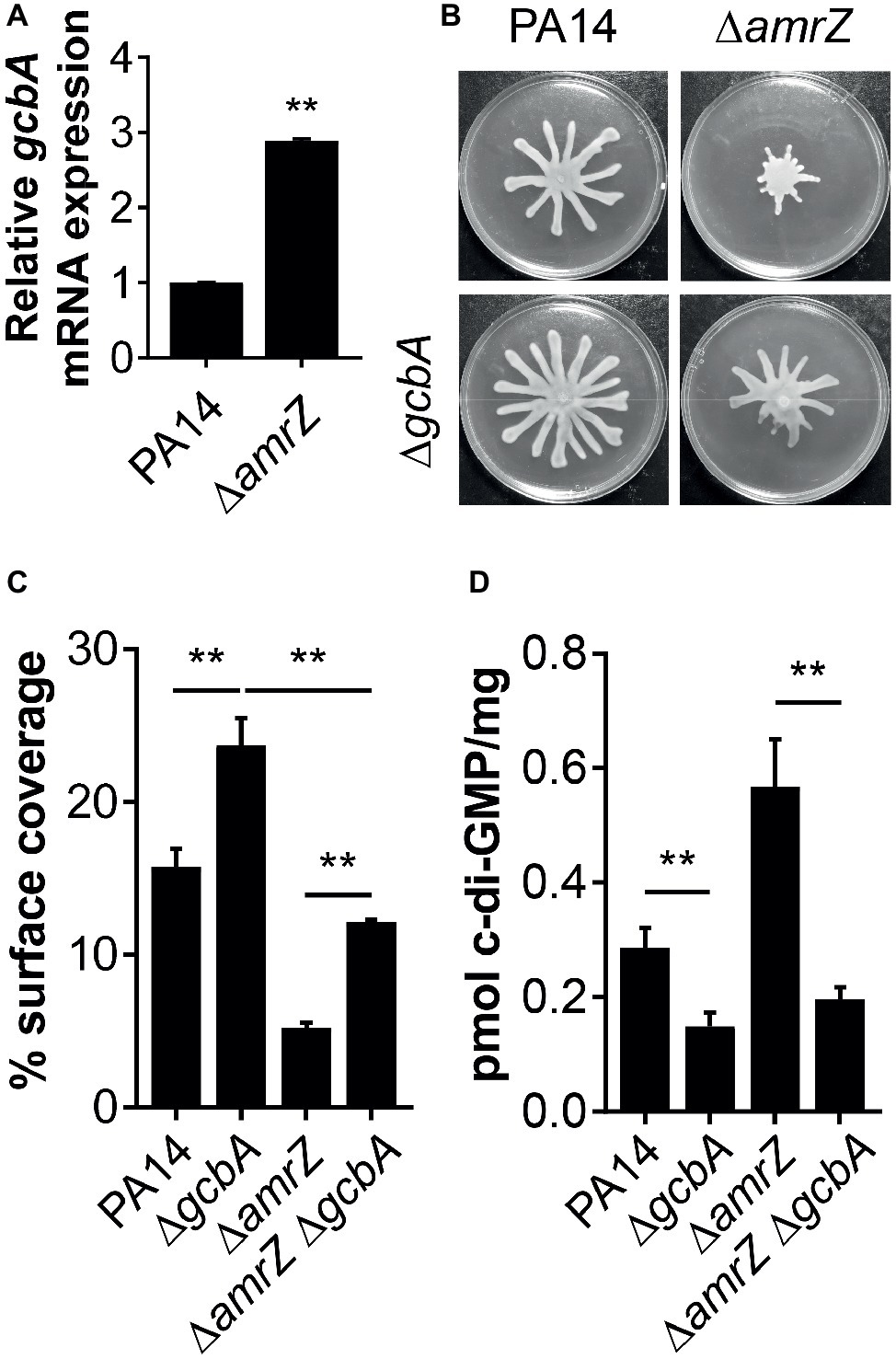
Figure 4. GcbA is required for increased c-di-GMP accumulation in ΔamrZ and partially responsible for its swarming phenotype. (A) Relative mRNA levels of gcbA in indicated strains under swarming conditions as determined by real-time PCR. (B) Representative images of swarming motilities of indicated strains. (C) Percent coverage of swarm plates by the respective swarms. (D) Quantification of c-di-GMP levels of the indicated strains grown on swarm plates. **p < 0.01 as determined by Student’s t-test.
FlgZ and PA14_56180 Are the Effector Relay Proteins That Respond to GcbA Cyclic di-GMP Signaling to Mediate Repression of Swarming Motility
We next set out to identify the downstream target(s) of GcbA c-di-GMP signaling, and expected certain c-di-GMP-binding effectors would likely be involved in this response. The P. aeruginosa genome encodes eight PilZ domain proteins, and seven of them are able to bind to c-di-GMP (Merighi et al., 2007). The seven PilZ domain proteins include HapZ (PA14_27930), FlgZ (PA14_20700), Alg44 (PA14_18550), and MapZ (PA14_60970) with known physiological roles and three other proteins (PA14_00130, PA14_25420, and PA14_56180) with unknown function. The PilZ (PA14_25770) shows no detectable binding of c-di-GMP in an in vitro assay but may interact with c-di-GMP in vivo (Merighi et al., 2007). To investigate whether PilZ domain proteins participate in the GcbA c-di-GMP mediated repression of swarming, we constructed deletions of each of the PilZ domain protein-encoding gene in the WT background, and the gcbA gene carried on a multi-copy plasmid (pUCP-gcbA) was introduced into these strains. The data revealed that overexpression of gcbA in WT PA14 completely abolished swarming motility compared with the vector control (Figure 5A, first two panels). Furthermore, overexpression of gcbA in most of the mutants lacking PilZ-domain bearing proteins, repressed swarming. The exceptions were the flgZ and PA14_56180 mutant strains. This strongly suggests that these genes are genetically linked to gcbA in controlling swarming motility in P. aeruginosa.
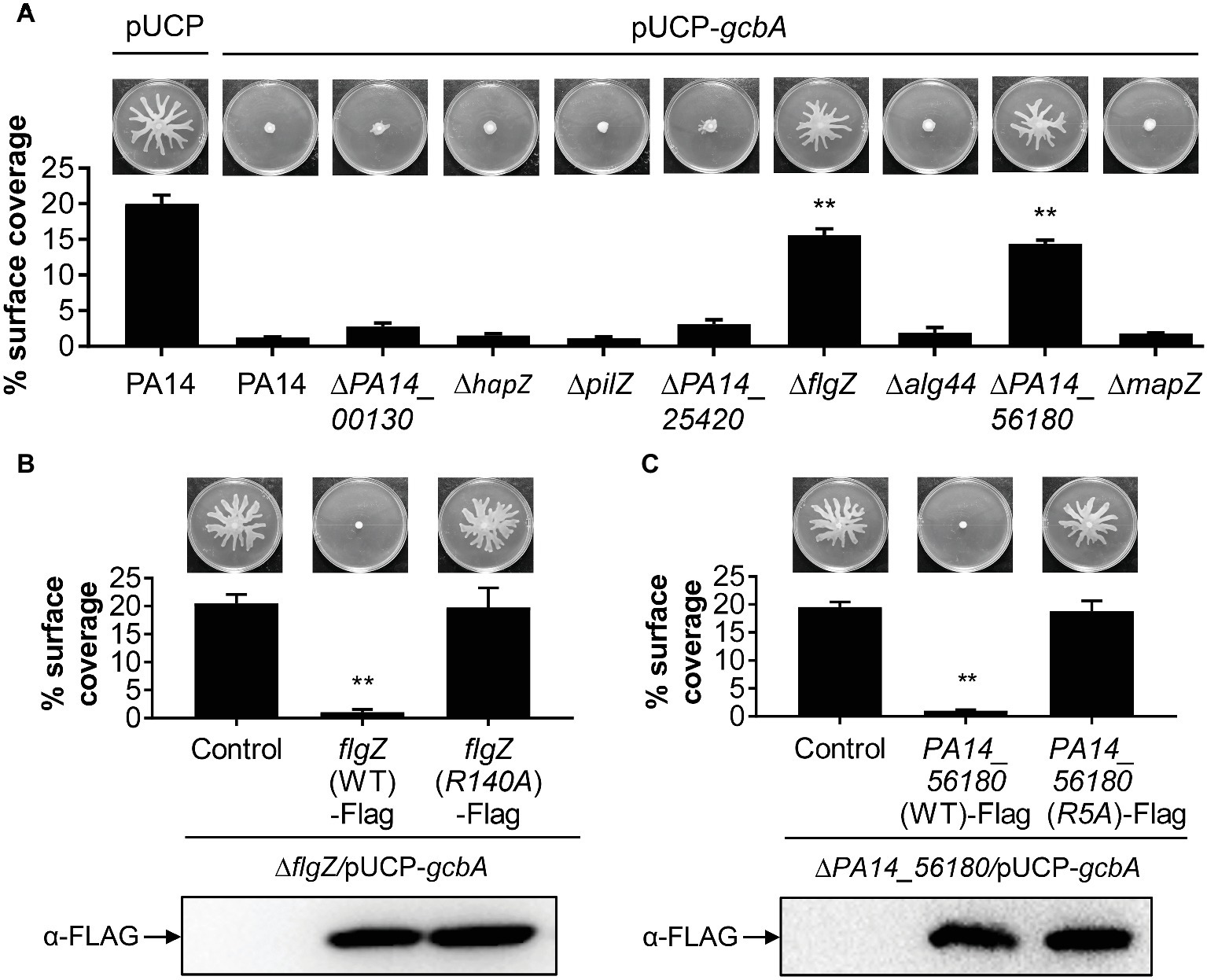
Figure 5. FlgZ and PA14_56180 are required for GcbA c-di-GMP-mediated swarming repression. (A) Swarming phenotype and quantification of the percentages of plate surface coverage of the indicated strains carrying either the empty vector (pUCP) or the GcbA overproducing plasmid pUCP-gcbA. **p < 0.01 (Student’s t-test, compared to PA14/pUCP-gcbA). (B) Representative swarming plates (upper row) and average percentage of the plate coverage (middle row) by ΔflgZ/pUCP-gcbA carrying the chromosomally encoded C-terminally FLAG-tagged WT FlgZ or FlgZ (R140A) under the control of the PBAD promoter at the attTn7 site. Significance was determined by Student’s t-test (**p < 0.01) relative to the vector control. Lower row: Western analysis of the FLAG-tagged FlgZ derivatives in indicated strains. Samples from equivalent number of bacterial cells were loaded onto SDS-PAGE gels and probed with anti-FLAG antibody. (C) Swarm analysis (upper row), quantification of percent coverage of swarm plates (middle row), and protein expression levels of FLAG-tagged derivatives of PA14_56180 (lower row) in ΔPA14_56180/pUCP-gcbA harboring the chromosomally encoded C-terminally FLAG-tagged WT PA14_56180 or PA14_56180 (R5A) as under panel (B).
PilZ domain proteins bind c-di-GMP by the conserved motifs RXXXR and (D/N)XSXXG (Cheang et al., 2019). Sequence alignment suggests that FlgZ and PA14_56180 contain both of these motifs (Supplementary Figure S2). In Pseudomonas putida, amino acid substitution analysis of FlgZ showed that alanine substitution of the first arginine in the RXXXR motif results in complete loss of c-di-GMP binding (Ko et al., 2010). Similarly, a corresponding replacement (R140A) of P. aeruginosa FlgZ abolishes the capacity of FlgZ to respond to c-di-GMP (Baker et al., 2016). Sequence alignment with orthologs from other species suggests that PA14_56180 is likely to use the same set of key residues (Supplementary Figure S2) for c-di-GMP binding (Xu et al., 2016a). To further validate whether c-di-GMP binding is required for FlgZ or PA14_56180 to mediate GcbA regulation of swarming, we generated C-terminally FLAG-tagged FlgZ-R140A and PA14_56180-R5A derivatives carrying amino acid substitution of the first arginine residue (R) in the RXXXR motif and assessed whether these mutant proteins were functional in swarming repression. As shown in Figures 5B,C, neither of these mutant variants mediated a reduced swarming phenotype in the GcbA overproducing background, even though they were expressed at equivalent levels as the WT protein, which readily repressed swarming in this background (Figures 5B,C). These data suggest that the conserved residue required for c-di-GMP binding is critical for FlgZ and PA14_56180 to mediate swarming repression in response to GcbA c-di-GMP signaling.
Furthermore, we asked whether expression of flgZ and PA14_56180 was affected by GcbA under the conditions studied here. qRT-PCR revealed that no significant differences in the transcript abundance of flgZ (fold change, 0.95 ± 0.05, for ΔgcbA versus PA14) or PA14_56180 (fold change, 0.83 ± 0.04) was noted between the ΔgcbA mutant and WT PA14 strain. Overexpression of gcbA similarly had no effect on expression of flgZ (fold change, 0.89 ± 0.03, for PA14/pUCP-gcbA versus PA14/pUCP) or PA14_56180 (fold change, 1.08 ± 0.06), suggesting that GcbA does not affect the transcription of flgZ and PA14_56180. Collectively, these results indicate that FlgZ and PA14_56180 function as the GcbA c-di-GMP effectors to mediate swarming repression in P. aeruginosa.
AmrZ Regulates Swarming Motility Through the GcbA-FlgZ/PA14_56180 Signal Transduction Pathway
Since GcbA contributes to the swarming repression in ΔamrZ, and FlgZ and PA14_56180 participate in the GcbA-mediated repression of swarming, we reasoned that if the negative regulation of swarming by GcbA occurred through the FlgZ/PA14_56180 pathway in ΔamrZ, deleting flgZ or PA14_56180 would be able to restore swarming to the ΔamrZ mutant. Therefore, we constructed mutations in either flgZ or PA14_56180 or both in the ΔamrZ mutant background as well as in WT as a control. As shown in Figures 6A,B, while deletion of either of the flgZ and PA14_56180 genes in ΔamrZ resulted in significantly enhanced motility compared to the ΔamrZ mutant, the ΔamrZ ΔflgZ ΔPA14_56180 triple mutant showed a more pronounced swarming phenotype than the individual ΔamrZ ΔflgZ or ΔamrZ ΔPA14_56180 mutants. However, the ΔamrZ ΔflgZ ΔPA14_56180 triple mutant still had a weaker swarming phenotype than the ΔflgZ ΔPA14_56180 double mutant, indicating that other factors are also involved in AmrZ-mediated swarming repression. Furthermore, we examined the expression of the two genes in ΔamrZ by qRT-PCR. Consistent with the previous RNA-Seq reports in PAO1 (Jones et al., 2014), we found the transcript abundances of flgZ and PA14_56180 in ΔamrZ were significantly increased (2.23 ± 0.06 and 2.42 ± 0.04, respectively) relative to the WT strain; together with the increased transcript levels of gcbA in ΔamrZ (Figure 4A), these results suggest a coordinated transcriptional regulation of gcbA, flgZ and PA14_56180 by AmrZ. Overall, these data suggest that swarming motility inhibition in the amrZ mutant is in part controlled by a c-di-GMP signaling module that consists of the DGC GcbA and two c-di-GMP receptors containing PilZ domains: FlgZ and PA14_56180.
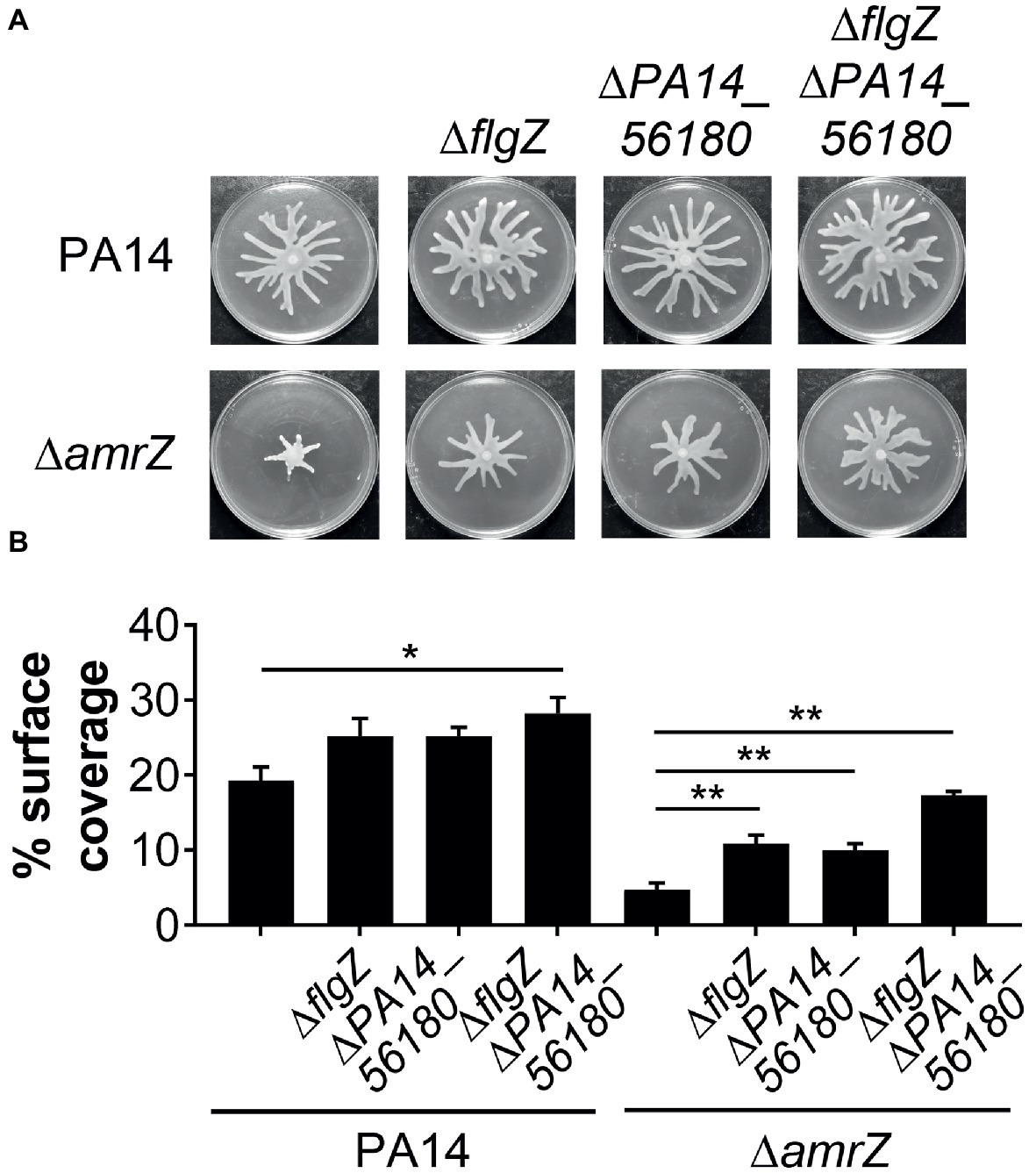
Figure 6. FlgZ and PA14_56180 contribute to swarming motility repression of the amrZ mutant. (A) Representative swarm plates of the indicated strains. (B) Percentage of the plate surface occupied by the respective swarms shown in (A). Significance was determined using one-way ANOVA followed by Tukey’s multiple comparison test (*p < 0.05; **p < 0.01).
Pel Exopolysaccharide Acts Additively With GcbA for Swarming Regulation in ΔamrZ
The epistasis studies with the amrZ and gcbA/flgZ/PA14_56180 genes described above indicated that eliminating GcbA signaling pathway in the ΔamrZ mutant background results in a significant, but not completely restoration of swarming motility (Figures 4B,C, 6A,B). Thus, there should be some other factor(s) responsible for the swarming defect in the ΔamrZ mutant. In PA14, exopolysaccharide (EPS) has been shown to be a regulator of swarming motility (Caiazza et al., 2007). We noticed that the amrZ mutant displayed an aggregated colony morphology when grown on agar plates, which is usually linked to overproduction of EPS (Kirisits et al., 2005). When tested on Congo red (CR) plates, the amrZ mutant indeed showed hyper-binding to CR and displayed a more wrinkly phenotype compared to the WT (Figure 7A; Supplementary Figure S3A), suggesting the amrZ mutant produced substantially more polysaccharides. The primary polysaccharide in PA14 is synthesized by the pel operon and this is the cellular component bound by CR (Friedman and Kolter, 2004; Lee et al., 2007). To investigate whether enhanced polysaccharide production contributes to the swarming repression in ΔamrZ, we introduced a pelA mutation into the ΔamrZ strain and examined its EPS production and swarming phenotype. The results showed that the pelA mutation eliminates both the CR binding and the wrinkled phenotype of the amrZ mutant (Figure 7A; Supplementary Figure S3A), and swarming motility was largely restored in the ΔamrZ ΔpelA mutant (Figures 7A,B), indicating that overproduction of Pel polysaccharide is responsible for the swarming defect observed in the amrZ mutant.
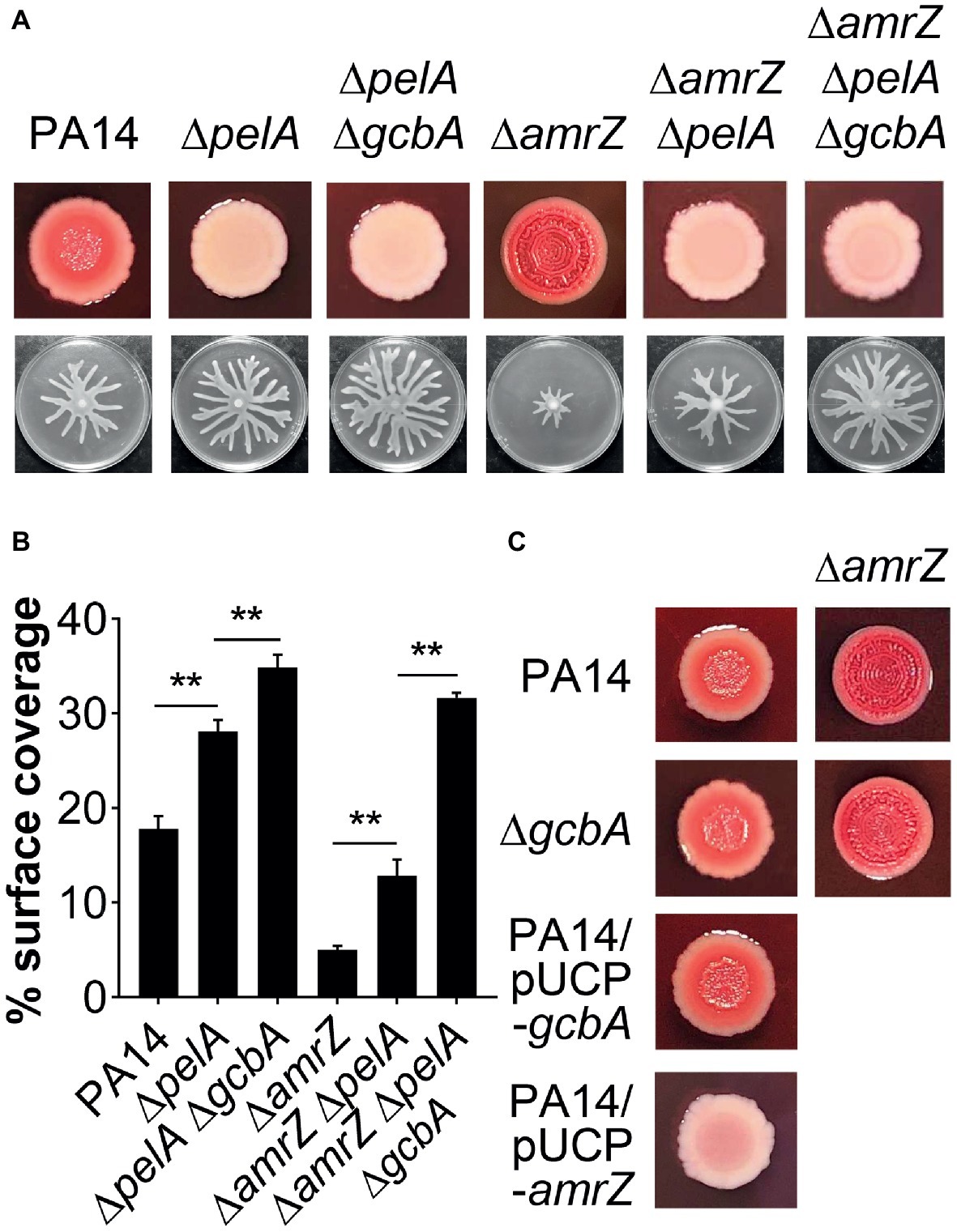
Figure 7. The pel-derived polysaccharide collaborates with GcbA to contribute to the swarming defect of ΔamrZ. (A) CR binding (upper row) and representative swarm plates (lower row) of the indicated strains. (B) Quantification of the extent of swarming for the strains indicated in (A). (C) CR-binding phenotypes of the indicated strains. The name of the corresponding strain is indicated on the left. In PA14 and ΔgcbA additional amrZ mutation was introduced as indicated at the top of the second column. Significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (**p < 0.01).
Regulation of Pel expression is complex. To assess whether increased Pel polysaccharide production in ΔamrZ is due to overexpression of the pel genes, we measured the transcript levels of the first (pelA) and last gene (pelG) of the pel operon (Friedman and Kolter, 2004) using qRT-PCR. Interestingly, we found that in cells grown on swarming plates, there was no significant change in pelA (fold change, 1.09 ± 0.04, for ΔamrZ versus PA14) or pelG (fold change, 1.15 ± 0.01) expression between the WT and ΔamrZ mutant. Similar results were observed when these strains were grown on CR plates under conditions identical to those used for CR assays (data not shown). These data suggest that the impact of AmrZ on Pel polysaccharide production in PA14 occurs via a nontranscriptional mechanism.
To determine whether GcbA c-di-GMP signaling crosstalks to the EPS production in ΔamrZ, we tested the CR binding ability of the ΔamrZ ΔgcbA mutant with the ΔamrZ strain. Interestingly, we observed that the ΔamrZ ΔgcbA mutant exhibited a CR binding phenotype comparable to that of the ΔamrZ mutant (Figure 7C; Supplementary Figure S3B). As a control, inactivation or overexpression of gcbA in WT PA14 also had no effect on CR binding (Figure 7C; Supplementary Figure S3B), which is consistent with a previous report that GcbA does not affect the transcription of pel genes (Petrova et al., 2014). Thus, these findings indicate that AmrZ regulation of EPS production is independent of GcbA c-di-GMP signaling. Consistent with this, inactivation of gcbA in the ΔamrZ ΔpelA mutant revealed additive effects on restoring the swarming defect of the amrZ mutant (Figures 7A,B), and the double ΔpelA ΔgcbA mutant also exhibited a significant increase of swarming relative to that observed for the ΔpelA mutant (Figures 7A,B). Together, these data suggest that Pel polysaccharide and GcbA act in concert to contribute to the repression of swarming in ΔamrZ.
Discussion
During colonization and infection, pathogenic bacteria such as P. aeruginosa has evolved several mechanisms to rapidly adjust gene expression to enable appropriate physiological and molecular adaptations. The transcription factor AmrZ is one of these regulators that has been implicated in the controlling of multiple cellular processes associated with virulence and environmental fitness (Jones et al., 2014; Martinez-Granero et al., 2014). Here, we are adding an important piece to the AmrZ regulon by showing that AmrZ is an important contributor to the regulation of surface-based swarming motility in P. aeruginosa. We have also demonstrated that the shifts in amrZ gene expression is associated with lifestyle changes, wherein actively moving cells express higher relative level of amrZ while a downregulation is observed in the biofilm growth mode. Combined with the fact that inactivation of amrZ results in enhanced level of biofilm formation (Jones et al., 2013), our results highlight the concept that AmrZ may serve as a molecular switch that controls the transition between motile and sessile lifestyles.
In agreement with the role of AmrZ in positive control of flagellum-driven motility in this study, recent findings have suggested that AmrZ functions as a positive regulator of motility in Pseudomonas syringae (Prada-Ramirez et al., 2016) and Pseudomonas stutzeri (Baltrus et al., 2018). In P. syringae, AmrZ activates the expression of the flagellin gene fliC and the amrZ mutant produces less flagella than the parental strain (Prada-Ramirez et al., 2016). This is in contrast to P. aeruginosa, where inactivation of amrZ does not affect the flagellin production (Baynham et al., 2006) or flagellation on the cell surface (Figure 2C). In P. stutzeri, AmrZ has also been shown to positively regulate colony spreading, a type of bacterial motility that is phenotypically presumed to be correlated with swarming motility (Baltrus et al., 2018). Interestingly, the P. aeruginosa AmrZ is able to complement both the swimming and colony spreading defects of the P. stutzeri amrZ mutant (Baltrus et al., 2018), suggesting that the two versions of this protein likely regulate motility in a similar way. However, the molecular mechanisms of motility regulation by AmrZ in P. stutzeri have not been determined.
Our work reveals that the second messenger c-di-GMP produced by the DGC GcbA is involved in the AmrZ-mediated regulation of swarming. c-di-GMP has long been known as a regulatory maestro that is involved in regulation of many cellular processes, including flagellum-based motility (Wolfe and Visick, 2008). GcbA was previously found to be an enzymatically active DGC participating in the regulation of motility in Pseudomonas fluorescens (Newell et al., 2011) and P. aeruginosa (Petrova et al., 2014). While the P. fluorescens GcbA inhibits swimming motility by a yet undetermined mechanism (Newell et al., 2011), P. aeruginosa GcbA was found to negatively regulate flagellum-driven motility by suppressing flagellar reversal rates (Petrova et al., 2014). Furthermore, the regulation of motility by GcbA is partially dependent on a small RNA (sRNA) RsmZ, and GcbA levels were found to positively correlate with the abundance of RsmZ (Petrova et al., 2014). In P. aeruginosa, transcription of rsmZ is under the direct control of the GacS/GacA two-component system (Brencic et al., 2009). However, there is no evidence of AmrZ regulating any of the members of the Gac/Rsm signaling cascade (Jones et al., 2014), and thus, other regulatory elements associated with GcbA c-di-GMP signaling might come into action to regulate motility in ΔamrZ.
In the past decades, the mechanisms of c-di-GMP turnover have been extensively studied (Schirmer and Jenal, 2009), yet the signal transduction mechanism such as the receptor(s) involved in specific DGCs/PDEs-associated c-di-GMP sensing still remain largely unknown (Krasteva et al., 2012). In this study, we provide evidence suggesting that two PilZ domain proteins FlgZ and PA14_56180 directly sense GcbA c-di-GMP signaling and act in concert to contribute to the repression of swarming motility in ΔamrZ. The flgZ gene is located downstream of the flgMN genes in a flagellar operon and encodes a homologue to YcgR (Baker et al., 2016), a c-di-GMP-binding protein in E. coli and Salmonella that acts as a flagellar brake to mediate c-di-GMP responsive control of motility (Paul et al., 2010). In P. aeruginosa, Baker et al. demonstrated that FlgZ interacts specifically with the flagellar stator protein MotC in a c-di-GMP-dependent manner, and absence of FlgZ suppressed the effect of the absence of a PDE (bifA) with regards to repression of swarming (Baker et al., 2016). Here, we found that FlgZ also contributes to GcbA c-di-GMP-mediated swarming repression (Figure 5), making GcbA a new DGC associated with the c-di-GMP receptor FlgZ. In addition, our study also identified PA14_56180 as being required for GcbA-mediated repression of swarming motility (Figure 5). Nevertheless, Baker and colleagues did not find PA14_56180, but only FlgZ as a regulator of swarming in the response to elevated c-di-GMP levels due to the absence of bifA (Baker et al., 2016). This suggests that PA14_56180 may specifically respond to GcbA c-di-GMP signaling while FlgZ may respond to a global pool of c-di-GMP. However, to understand the specificity and generality of these receptors in c-di-GMP signal transmission will require the identification of individual c-di-GMP signaling pathway of the multiple DGCs and PDEs encoded in P. aeruginosa genome. In another study, Lewis and colleagues performed a screen for proteins involved in c-di-GMP-mediated response to ethanol which led to the identification of a few candidates including GcbA and FlgZ (Lewis et al., 2019); however, the relationship between the two proteins was not investigated in this research, and our findings may provide an explanation for this screening result. Unlike FlgZ, there is limited information available about PA14_56180, but our results highlight a role for PA14_56180 as a GcbA c-di-GMP receptor affecting swarming motility of P. aeruginosa. Besides, the expression of gcbA, flgZ and PA14_56180 are all upregulated in the amrZ mutant, suggesting that these genes are cotranscriptionally regulated. Furthermore, the participation of PA14_56180 in the GcbA pathway may hint at potential for colocalization of GcbA and PA14_56180 within the cell. However, it is also possible that other c-di-GMP signaling could feed in to modulate swarming motility through PA14_56180. Future studies will be required to determine PA14_56180 subcellular localization and the molecular basis for downstream signaling, including the impact of c-di-GMP binding on the structure and function of PA14_56180. Elucidating such questions will substantially enhance our understanding of c-di-GMP signaling mechanisms.
In addition to the GcbA-FlgZ/PA14_56180 signal transduction cascade, our data suggest that AmrZ can modulate another facet of swarming by coordinating the production of Pel polysaccharide. We show that the amrZ mutant produces more EPS and overexpression of amrZ in WT leads to repression of EPS production (Figures 7A,C; Supplementary Figure S3B). However, the ΔamrZ mutation does not alter pel mRNA expression, suggesting that AmrZ impacts Pel polysaccharide via a nontranscriptional mechanism. This is distinct from the RNA-seq data in PAO1, where AmrZ activates expression of the pel polysaccharide operon (Jones et al., 2014). We also confirmed that a slight decrease in CR binding was observed for a PAO1 amrZ mutant (RCRB value: 0.07 ± 0.01) compared to that of the WT PAO1 (0.12 ± 0.01). Indeed, PAO1 differs from PA14 in that PAO1 uses Psl as the primary matrix polysaccharide, while PA14 has a three-gene deletion in the psl operon and uses Pel (Colvin et al., 2012; Visaggio et al., 2015). One possible explanation for this is the differences in experimental conditions where some other regulatory elements might come into action to affect the function of AmrZ in transcriptional regulation, or some yet-to-be discovered post-transcriptional mechanisms maybe involved in the AmrZ-mediated regulation of Pel production in PA14. A possible candidate for this post-transcriptional regulation is the AlgC protein which is a key enzyme that provides sugar precursors for the synthesis of P. aeruginosa exopolysaccharides including Pel (Ma et al., 2012). Whether the expression or activity of AlgC is altered in ΔamrZ and the role of AlgC in regulation of the synthesis of Pel polysaccharide in PA14 will be the subject of future studies. Furthermore, our results suggest that the Pel polysaccharide acts in concert with GcbA signal to contribute to the swarming repression in ΔamrZ, as combinatorial analysis of them revealed additive effects on swarming. Similar collaboration has also been found in Salmonella, whereby the EPS cellulose works in cooperation with the c-di-GMP-binding protein YcgR to inhibit flagellar motility under high c-di-GMP conditions (Zorraquino et al., 2013). In the P. aeruginosa ΔbifA and ΔhptB mutants that exhibit elevated levels of c-di-GMP, Pel polysaccharide and FlgZ also function together to mediate c-di-GMP-dependent repression of swarming (Baker et al., 2016). However, including the AmrZ-dependent mechanism reported here, we cannot rule out the possibility that Pel polysaccharide and the downstream mediators (such as FlgZ, PA14_56180, etc.) of the c-di-GMP signaling pathway may influence each other. This issue will be elucidated in future studies.
Our findings are summarized in a model shown in Figure 8. We propose that P. aeruginosa adjusts the expression of amrZ in response to proper environmental signals such as nutritions or changes in medium viscosity during surface growth. Through AmrZ, P. aeruginosa could modulate swarming motility by negatively regulating Pel polysaccharide production and a GcbA-dependent c-di-GMP signaling. While the Pel polysaccharide contributes to the progression toward irreversible attachment (Caiazza et al., 2007), the GcbA c-di-GMP signal is then transmitted via FlgZ to the flagellar stator MotC, resulting in impairment of flagellar function (Baker et al., 2016). The GcbA signal can also be sensed by another c-di-GMP receptor PA14_56180, which impacts swarming through an as-yet-unknown mechanism. In view of the importance of bacterial motility for their survival in a natural environment and during infection inside a host, with such a double checkpoint mechanism, AmrZ would allow P. aeruginosa to delicately control its surface-associated behaviors, thus enabling better adaptations in response to environmental fluctuations.
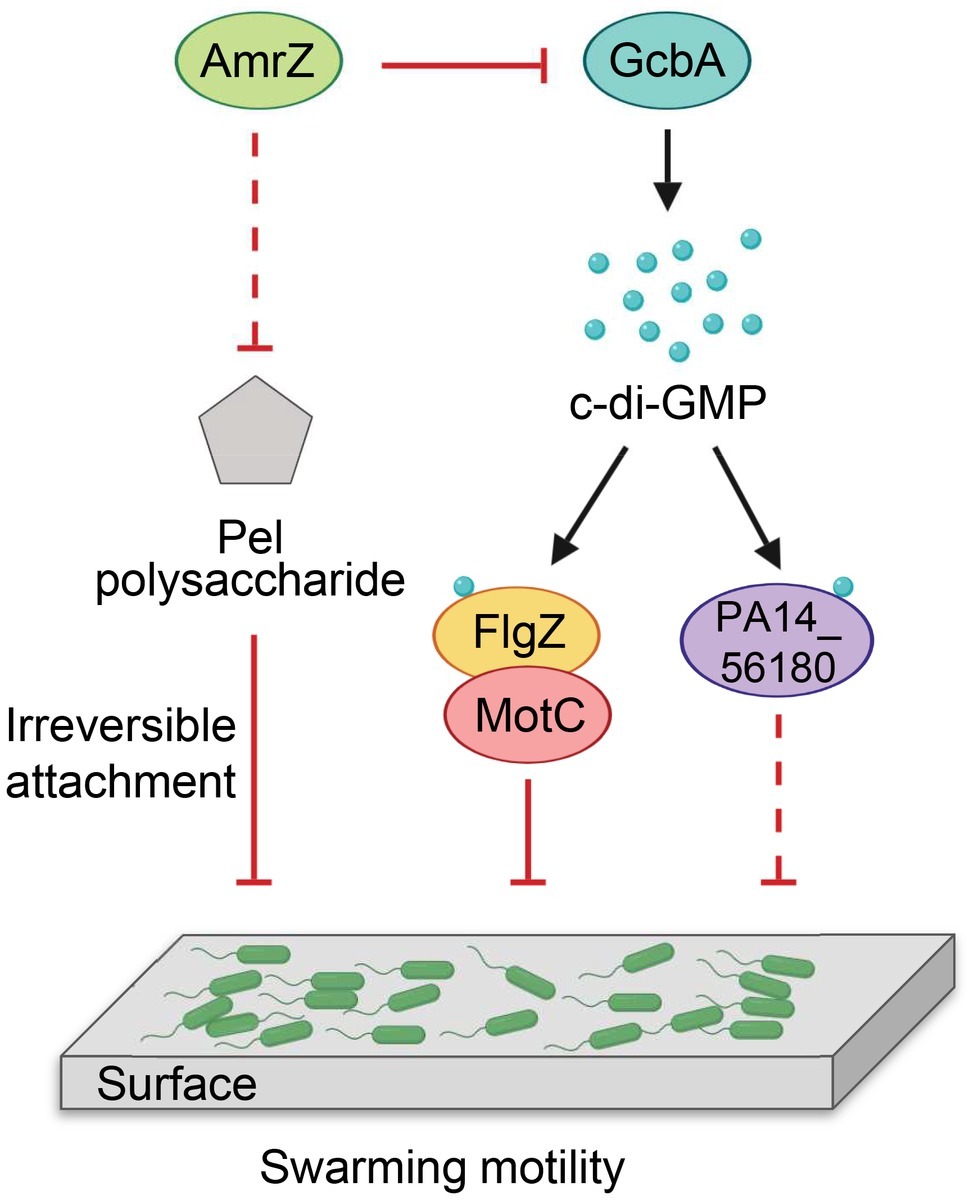
Figure 8. Model for AmrZ-mediated swarming motility control. AmrZ affects swarming by negatively regulating the production of Pel polysaccharide and a GcbA-dependent c-di-GMP signaling. The Pel polysaccharide inhibits swarming motility by promoting the transition from reversible to irreversible attachment (Caiazza et al., 2007). The GcbA c-di-GMP signal can be further sensed by two PilZ-containing proteins, FlgZ and PA14_56180. Binding of c-di-GMP to FlgZ induces its interaction with the MotC stator and impairs flagellar function probably by preventing the engagement of MotCD with the rotor (Baker et al., 2016). PA14_56180 regulates swarming in a c-di-GMP-dependent manner, but the downstream target(s) of PA14_56180 remains unknown. The solid lines represent direct regulations, and the dashed lines indicate probable indirect regulations.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
KL, QB, and LH conceived and designed the experiments. LH, AD, and QC performed the experiments. LH, KL, and QB analyzed the data. LH and KL wrote the paper.
Funding
This work was supported by the National Natural Science Foundation of China (31500109, 81870075), the Natural Science Foundation for Youths of Zhejiang Province (LQ16C010001), China Scholarship Council (CSC) Grant (201708330005) and a startup fund to KL from Wenzhou Medical University (QTJ14024).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank George A. O’Toole, Sherry L. Kuchma and Shanice S. Webster for providing technical assistance in the swarming motility assay, Zhouxi Fang for help and advice with the electron microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01847/full#supplementary-material
References
Allsopp, L. P., Wood, T. E., Howard, S. A., Maggiorelli, F., Nolan, L. M., Wettstadt, S., et al. (2017). RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 114, 7707–7712. doi: 10.1073/pnas.1700286114
Amikam, D., and Galperin, M. Y. (2006). PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6. doi: 10.1093/bioinformatics/bti739
Andrews, G. P., and Maurelli, A. T. (1992). mxiA of Shigella flexneri 2a, which facilitates export of invasion plasmid antigens, encodes a homolog of the low-calcium-response protein, LcrD, of Yersinia pestis. Infect. Immun. 60, 3287–3295. doi: 10.1007/BF01989988
Baker, A. E., Diepold, A., Kuchma, S. L., Scott, J. E., Ha, D. G., Orazi, G., et al. (2016). PilZ domain protein FlgZ mediates cyclic di-GMP-dependent swarming motility control in Pseudomonas aeruginosa. J. Bacteriol. 198, 1837–1846. doi: 10.1128/JB.00196-16
Baker, A. E., Webster, S. S., Diepold, A., Kuchma, S. L., Bordeleau, E., Armitage, J. P., et al. (2019). Flagellar stators stimulate c-di-GMP production by Pseudomonas aeruginosa. J. Bacteriol. [Preprint]. doi: 10.1128/JB.00741-18
Baltrus, D. A., Dougherty, K., Diaz, B., and Murillo, R. (2018). Evolutionary plasticity of AmrZ regulation in Pseudomonas. mSphere 3:e00132. doi: 10.1128/mSphere.00132-18
Baynham, P. J., Ramsey, D. M., Gvozdyev, B. V., Cordonnier, E. M., and Wozniak, D. J. (2006). The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188, 132–140. doi: 10.1128/JB.188.1.132-140.2006
Baynham, P. J., and Wozniak, D. J. (1996). Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22, 97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Caiazza, N. C., Merritt, J. H., Brothers, K. M., and O’Toole, G. A. (2007). Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 3603–3612. doi: 10.1128/JB.01685-06
Caiazza, N. C., Shanks, R. M., and O’Toole, G. A. (2005). Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187, 7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005
Cangelosi, G. A., Palermo, C. O., Laurent, J. P., Hamlin, A. M., and Brabant, W. H. (1999). Colony morphotypes on Congo red agar segregate along species and drug susceptibility lines in the Mycobacterium avium-intracellulare complex. Microbiology 145, 1317–1324. doi: 10.1099/13500872-145-6-1317
Cheang, Q. W., Xin, L., Chea, R. Y. F., and Liang, Z. X. (2019). Emerging paradigms for PilZ domain-mediated C-di-GMP signaling. Biochem. Soc. Trans. 47, 381–388. doi: 10.1042/BST20180543
Chen, A. I., Dolben, E. F., Okegbe, C., Harty, C. E., Golub, Y., Thao, S., et al. (2014). Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog. 10:e1004480. doi: 10.1371/journal.ppat.1004480
Choi, K.-H., Kumar, A., and Schweizer, H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397. doi: 10.1016/j.mimet.2005.06.001
Choi, K. H., and Schweizer, H. P. (2006). Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161. doi: 10.1038/nprot.2006.24
Colvin, K. M., Irie, Y., Tart, C. S., Urbano, R., Whitney, J. C., Ryder, C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x
Costaglioli, P., Barthe, C., Fayon, M., Christoflour, N., Bui, S., Derlich, L., et al. (2014). Selection of Pseudomonas aeruginosa reference genes for RT-qPCR analysis from sputum of cystic fibrosis patients. Mol. Cell. Probes 28, 10–12. doi: 10.1016/j.mcp.2013.09.003
Deziel, E., Lepine, F., Milot, S., and Villemur, R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149, 2005–2013. doi: 10.1099/mic.0.26154-0
Frangipani, E., Visaggio, D., Heeb, S., Kaever, V., Camara, M., Visca, P., et al. (2014). The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 16, 676–688. doi: 10.1111/1462-2920.12164
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Ghorbal, S. K. B., Chourabi, K., Maalej, L., Ammar, A. B., Ouzari, H. I., Hassen, A., et al. (2019). Pseudomonas aeruginosa swarmer cells adaptation toward UVc radiations. Front. Microbiol. 10:556. doi: 10.3389/fmicb.2019.00556
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Ha, D. G., Kuchma, S. L., and O’Toole, G. A. (2014). Plate-based assay for swarming motility in Pseudomonas aeruginosa. Methods Mol. Biol. 1149, 67–72. doi: 10.1007/978-1-4939-0473-0_8
Ha, D. G., and O’Toole, G. A. (2015). C-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. 3:MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Jean-Pierre, F., Tremblay, J., and Deziel, E. (2016). Broth versus surface-grown cells: differential regulation of RsmY/Z small RNAs in Pseudomonas aeruginosa by the Gac/HptB system. Front. Microbiol. 7:2168. doi: 10.3389/fmicb.2016.02168
Jones, C. J., Newsom, D., Kelly, B., Irie, Y., Jennings, L. K., Xu, B., et al. (2014). ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10:e1003984. doi: 10.1371/journal.ppat.1003984
Jones, C. J., Ryder, C. R., Mann, E. E., and Wozniak, D. J. (2013). AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J. Bacteriol. 195, 1637–1644. doi: 10.1128/JB.02190-12
Kearns, D. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Kirisits, M. J., Prost, L., Starkey, M., and Parsek, M. R. (2005). Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71, 4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005
Ko, J., Ryu, K. S., Kim, H., Shin, J. S., Lee, J. O., Cheong, C., et al. (2010). Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by PilZ domain proteins. J. Mol. Biol. 398, 97–110. doi: 10.1016/j.jmb.2010.03.007
Kohler, T., Curty, L. K., Barja, F., van Delden, C., and Pechere, J. C. (2000). Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182, 5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000
Kong, W., Chen, L., Zhao, J., Shen, T., Surette, M. G., Shen, L., et al. (2013). Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 88, 784–797. doi: 10.1111/mmi.12223
Krasteva, P. V., Giglio, K. M., and Sondermann, H. (2012). Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci. 21, 929–948. doi: 10.1002/pro.2093
Kuchma, S. L., Brothers, K. M., Merritt, J. H., Liberati, N. T., Ausubel, F. M., and O’Toole, G. A. (2007). BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 8165–8178. doi: 10.1128/JB.00586-07
Kuchma, S. L., Delalez, N. J., Filkins, L. M., Snavely, E. A., Armitage, J. P., and O’Toole, G. A. (2015). Cyclic Di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J. Bacteriol. 197, 420–430. doi: 10.1128/JB.02130-14
Kuchma, S. L., Griffin, E. F., and O’Toole, G. A. (2012). Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J. Bacteriol. 194, 5388–5403. doi: 10.1128/JB.00899-12
Lee, V. T., Matewish, J. M., Kessler, J. L., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x
Lewis, K. A., Baker, A. E., Chen, A. I., Harty, C. E., Kuchma, S. L., O’Toole, G. A., et al. (2019). Ethanol decreases Pseudomonas aeruginosa flagellar motility through the regulation of flagellar stators. J. Bacteriol. [Preprint]. doi: 10.1128/JB.00285-19
Li, K., Xu, C., Jin, Y., Sun, Z., Liu, C., Shi, J., et al. (2013). SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. MBio 4, e00419–e00413. doi: 10.1128/mBio.00419-13
Li, K., Yang, G., Debru, A. B., Li, P., Zong, L., Li, P., et al. (2017). SuhB regulates the motile-sessile switch in Pseudomonas aeruginosa through the Gac/Rsm pathway and c-di-GMP signaling. Front. Microbiol. 8:1045. doi: 10.3389/fmicb.2017.01045
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L., Wang, J., Wang, S., Anderson, E. M., Lam, J. S., Parsek, M. R., et al. (2012). Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol. 14, 1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x
Martinez-Granero, F., Redondo-Nieto, M., Vesga, P., Martin, M., and Rivilla, R. (2014). AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genomics 15:237. doi: 10.1186/1471-2164-15-237
McCarter, L. L., and Gomelsky, M. (2015). Fifty ways to inhibit motility via cyclic Di-GMP: the emerging Pseudomonas aeruginosa swarming story. J. Bacteriol. 197, 406–409. doi: 10.1128/JB.02483-14
Merighi, M., Lee, V. T., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). The second messenger bis-(3’-5’)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895. doi: 10.1111/j.1365-2958.2007.05817.x
Merritt, J. H., Ha, D. G., Cowles, K. N., Lu, W., Morales, D. K., Rabinowitz, J., et al. (2010). Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. MBio 1, e00183–e00110. doi: 10.1128/mBio.00183-10
Moscoso, J. A., Mikkelsen, H., Heeb, S., Williams, P., and Filloux, A. (2011). The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x
Murray, T. S., and Kazmierczak, B. I. (2008). Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J. Bacteriol. 190, 2700–2708. doi: 10.1128/JB.01620-07
Murray, T. S., Ledizet, M., and Kazmierczak, B. I. (2010). Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J. Med. Microbiol. 59, 511–520. doi: 10.1099/jmm.0.017715-0
Newell, P. D., Yoshioka, S., Hvorecny, K. L., Monds, R. D., and O’Toole, G. A. (2011). Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J. Bacteriol. 193, 4685–4698. doi: 10.1128/JB.05483-11
Orr, M. W., and Lee, V. T. (2016). A PilZ domain protein for chemotaxis adds another layer to c-di-GMP-mediated regulation of flagellar motility. Sci. Signal. 9:fs16. doi: 10.1126/scisignal.aai8859
Overhage, J., Bains, M., Brazas, M. D., and Hancock, R. E. (2008). Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190, 2671–2679. doi: 10.1128/JB.01659-07
Partridge, J. D., and Harshey, R. M. (2013). Swarming: flexible roaming plans. J. Bacteriol. 195, 909–918. doi: 10.1128/JB.02063-12
Paul, K., Nieto, V., Carlquist, W. C., Blair, D. F., and Harshey, R. M. (2010). The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139. doi: 10.1016/j.molcel.2010.03.001
Petrova, O. E., Cherny, K. E., and Sauer, K. (2014). The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J. Bacteriol. 196, 2827–2841. doi: 10.1128/JB.01628-14
Prada-Ramirez, H. A., Perez-Mendoza, D., Felipe, A., Martinez-Granero, F., Rivilla, R., Sanjuan, J., et al. (2016). AmrZ regulates cellulose production in Pseudomonas syringae pv. Tomato DC3000. Mol. Microbiol. 99, 960–977. doi: 10.1111/mmi.13278
Price, K. E., Naimie, A. A., Griffin, E. F., Bay, C., and O’Toole, G. A. (2016). Tobramycin-treated Pseudomonas aeruginosa PA14 enhances Streptococcus constellatus 7155 biofilm formation in a cystic fibrosis model system. J. Bacteriol. 198, 237–247. doi: 10.1128/JB.00705-15
Pryor, E. E. Jr., Waligora, E. A., Xu, B., Dellos-Nolan, S., Wozniak, D. J., and Hollis, T. (2012). The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 8:e1002648. doi: 10.1371/journal.ppat.1002648
Rashid, M. H., and Kornberg, A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97, 4885–4890. doi: 10.1073/pnas.060030097
Romling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Schirmer, T., and Jenal, U. (2009). Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7, 724–735. doi: 10.1038/nrmicro2203
Siegmund, I., and Wagner, F. (1991). New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 5, 265–268. doi: 10.1007/BF02438660
Silby, M. W., Winstanley, C., Godfrey, S. A., Levy, S. B., and Jackson, R. W. (2011). Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680. doi: 10.1111/j.1574-6976.2011.00269.x
Spangler, C., Bohm, A., Jenal, U., Seifert, R., and Kaever, V. (2010). A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81, 226–231. doi: 10.1016/j.mimet.2010.03.020
Tart, A. H., Blanks, M. J., and Wozniak, D. J. (2006). The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 188, 6483–6489. doi: 10.1128/JB.00636-06
Toutain, C. M., Zegans, M. E., and O’Toole, G. A. (2005). Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187, 771–777. doi: 10.1128/JB.187.2.771-777.2005
Tremblay, J., and Deziel, E. (2010). Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics 11:587. doi: 10.1186/1471-2164-11-587
Valentini, M., and Filloux, A. (2016). Biofilms and cyclic di-GMP (c-di-GMP) Signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555. doi: 10.1074/jbc.R115.711507
Visaggio, D., Pasqua, M., Bonchi, C., Kaever, V., Visca, P., and Imperi, F. (2015). Cell aggregation promotes pyoverdine-dependent iron uptake and virulence in Pseudomonas aeruginosa. Front. Microbiol. 6:902. doi: 10.3389/fmicb.2015.00902
West, S., Schweizer, H., Dall, C., Sample, A., and Runyen-Janecky, L. (1994). Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86. doi: 10.1016/0378-1119(94)90237-2
Wolfe, A. J., and Visick, K. L. (2008). Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190, 463–475. doi: 10.1128/JB.01418-07
Xu, L., Venkataramani, P., Ding, Y., Liu, Y., Deng, Y., Yong, G. L., et al. (2016a). A cyclic di-GMP-binding adaptor protein interacts with histidine kinase to regulate two-component Signaling. J. Biol. Chem. 291, 16112–16123. doi: 10.1074/jbc.M116.730887
Xu, L., Xin, L., Zeng, Y., Yam, J. K., Ding, Y., Venkataramani, P., et al. (2016b). A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci. Signal. 9:ra102. doi: 10.1126/scisignal.aaf7584
Yan, J., Deforet, M., Boyle, K. E., Rahman, R., Liang, R., Okegbe, C., et al. (2017). Bow-tie signaling in c-di-GMP: machine learning in a simple biochemical network. PLoS Comput. Biol. 13:e1005677. doi: 10.1371/journal.pcbi.1005677
Yeung, A. T., Parayno, A., and Hancock, R. E. (2012). Mucin promotes rapid surface motility in Pseudomonas aeruginosa. MBio 3, e00073–e00012. doi: 10.1128/mBio.00073-12
Yeung, A. T., Torfs, E. C., Jamshidi, F., Bains, M., Wiegand, I., Hancock, R. E., et al. (2009). Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J. Bacteriol. 191, 5592–5602. doi: 10.1128/JB.00157-09
Keywords: Pseudomonas aeruginosa, AmrZ, swarming motility, cyclic di-GMP, GcbA, FlgZ, PA14_56180, exopolysaccharide
Citation: Hou L, Debru A, Chen Q, Bao Q and Li K (2019) AmrZ Regulates Swarming Motility Through Cyclic di-GMP-Dependent Motility Inhibition and Controlling Pel Polysaccharide Production in Pseudomonas aeruginosa PA14. Front. Microbiol. 10:1847. doi: 10.3389/fmicb.2019.01847
Edited by:
Anne Galinier, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
David Salvador Zamorano Sanchez, Universidad Nacional Autónoma de México, MexicoFrancesco Imperi, Roma Tre University, Italy
Copyright © 2019 Hou, Debru, Chen, Bao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kewei Li, curwaylee@wmu.edu.cn; curwaylee@aliyun.com; Qiyu Bao, baoqy@genomics.cn
 Lingli Hou
Lingli Hou Alexander Debru
Alexander Debru Qianqian Chen
Qianqian Chen Qiyu Bao
Qiyu Bao Kewei Li
Kewei Li