Current Strategies of Antiviral Drug Discovery for COVID-19
- 1Tsinghua-Peking Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, MOE Key Laboratory of Bioorganic Phosphorus Chemistry & Chemical Biology, School of Pharmaceutical Sciences, Center for infectious Disease Research, School of Medicine, Tsinghua University, Beijing, China
- 2Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, MOE Key Laboratory of Bioorganic Phosphorus Chemistry & Chemical Biology, School of Pharmaceutical Sciences, Center for Infectious Disease Research, School of Medicine, Tsinghua University, Beijing, China
SARS-CoV-2 belongs to the family of enveloped, single-strand RNA viruses known as Betacoronavirus in Coronaviridae, first reported late 2019 in China. It has since been circulating world-wide, causing the COVID-19 epidemic with high infectivity and fatality rates. As of the beginning of April 2021, pandemic SARS-CoV-2 has infected more than 130 million people and led to more than 2.84 million deaths. Given the severity of the epidemic, scientists from academia and industry are rushing to identify antiviral strategies to combat the disease. There are several strategies in antiviral drugs for coronaviruses including empirical testing of known antiviral drugs, large-scale phenotypic screening of compound libraries and target-based drug discovery. To date, an increasing number of drugs have been shown to have anti-coronavirus activities in vitro and in vivo, but only remdesivir and several neutralizing antibodies have been approved by the US FDA for treating COVID-19. However, remdesivir’s clinical effects are controversial and new antiviral drugs are still urgently needed. We will discuss the current status of the drug discovery efforts against COVID-19 and potential future directions. With the ever-increasing movability of human population and globalization of world economy, emerging and reemerging viral infectious diseases seriously threaten public health. Particularly the past and ongoing outbreaks of coronaviruses cause respiratory, enteric, hepatic and neurological diseases in infected animals and human (Woo et al., 2009). The human coronavirus (HCoV) strains (HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1) usually cause common cold with mild, self-limiting upper respiratory tract infections. By contrast, the emergence of three deadly human betacoronaviruses, middle east respiratory syndrome coronavirus (MERS) (Zaki et al., 2012), severe acute respiratory syndrome coronavirus (SARS-CoV) (Lee et al., 2003), the SARS-CoV-2 (Jin et al., 2020a) highlight the need to identify new treatment strategies for viral infections. SARS-CoV-2 is the etiological agent of COVID-19 disease named by World Health Organization (WHO) (Zhu N. et al., 2020). This disease manifests as either an asymptomatic infection or a mild to severe pneumonia. This pandemic disease causes extent morbidity and mortality in the whole world, especially regions out of China. Similar to SARS and MERS, the SARS CoV-2 genome encodes four structural proteins, sixteen non-structural proteins (nsp) and accessory proteins. The structural proteins include spike (S), envelope (E), membrane (M), nucleoprotein (N). The spike glycoprotein directly recognizes and engages cellular receptors during viral entry. The four non-structural proteins including papain-like protease (PLpro), 3-chymotrypsin-like protease (3CLpro), helicase, and RNA-dependent RNA polymerase (RdRp) are key enzymes involved in viral transcription and replication. The spike and the four key enzymes were considered attractive targets to develop antiviral agents (Zumla et al., 2016). The catalytic sites of the four enzymes of SARS-CoV2 share high similarities with SARS CoV and MERS in genomic sequences (Morse et al., 2020). Besides, the structures of the key drug-binding pockets are highly conserved among the three coronaviruses (Morse et al., 2020). Therefore, it follows naturally that existing anti-SARS-CoV and anti-MERS drugs targeting these enzymes can be repurposed for SARS-CoV-2. Based on previous studies in SARS-CoV and MERS-CoV, it is anticipated a number of therapeutics can be used to control or prevent emerging infectious disease COVID-19 (Li and de Clercq, 2020; Wang et al., 2020c; Ita, 2021), these include small-molecule drugs, peptides, and monoclonal antibodies. Given the urgency of the SARS-CoV-2 outbreak, here we discuss the discovery and development of new therapeutics for SARS-CoV-2 infection based on the strategies from which the new drugs are derived.
Empirically Repositioning of Existing Antiviral Drugs Against COVID-19
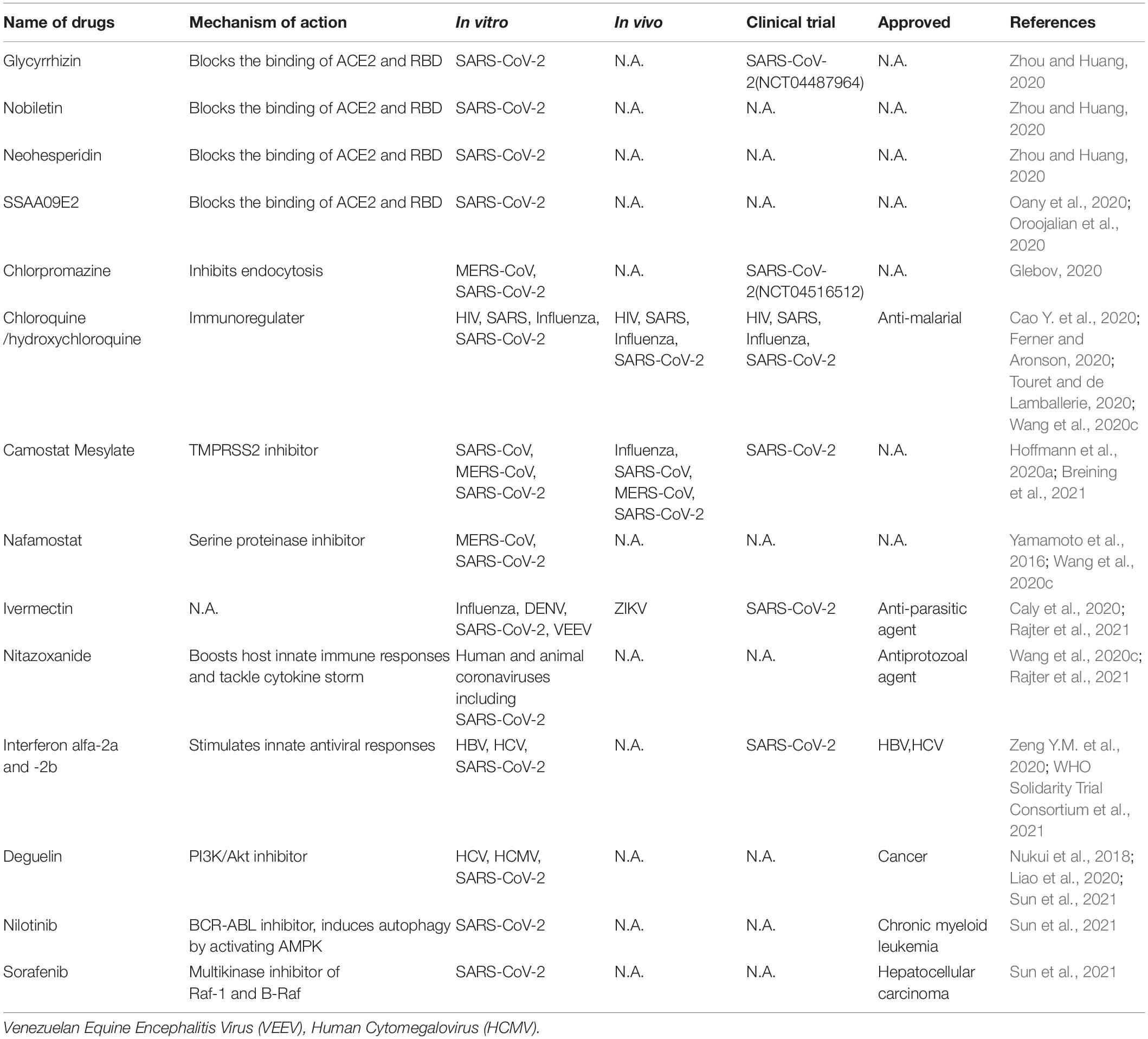
Currently, there is no highly efficacious and specific treatment for SARS-CoV-2. Therefore, it is urgent need to identify effective antiviral agents from existing drugs to combat the infection. These drugs can be rapidly repurposed to clinic application for treating COVID-19 patients given their proven safety. These compounds include direct acting antivirals (Table 1) and host targeting antiviral agents (Table 2).
Direct-Acting Antivirals
The functional domains of the spike protein of SARS-CoV-2 are highly conserved with those of SARS-CoV (Xia et al., 2020b). These domains can be classified into S1 (aa 14-685) and S2 (aa 686-1273) subunit. S1 subunit includes the N-terminal domain (NTD), the receptor-binding domain (RBD), and the receptor-binding motif (RBM), while the S2 subunit consists of the fusion peptides, heptad repeat regions (HR1, HR2), the transmembrane domain (TMD) and the cytoplasm domain (CD). HR1 and HR2, each having three alpha helical segments, pack together to form the six-helical bundle (6-HB). This 6-HB structure is similar to that of HIV envelope protein in the form as well as the function, which is to facilitate virus entry into the cells. Based on the development of HIV fusion peptide inhibitor targeting the 6-HB of HIV, EK1, a peptide-based fusion inhibitor, was developed to target the HR1 domain in divergent human coronaviruses. EK1C4, modified by adding PEG and cholesterol moieties to the EK1 peptide, more potently inhibited SARS-CoV-2 spike-mediated membrane fusion and pseudovirus infection (IC50 = 15.8 nM for inhibition of SARS-CoV-2 pseudovirus). Its inhibitory ability is highly effective against multiple coronaviruses including SARS-CoV and SARS-CoV-2 in human cells and mouse models with low or no toxicity to the hosts (Xia et al., 2020a), thereby representing a pan-coronavirus inhibitor that potentially applies to strain variants of SARS-CoV-2 and coronaviruses that might emerge in the future.
Arbidol, an approved anti-influenza drug, inhibits early stage of viral replication by blocking the virus fusion with the cell membrane, including SARS-CoV-2 (Wu et al., 2020a). It can effectively inhibit SARS-CoV-2 infection in vitro via impeding spike trimerization (Vankadari, 2020) and blocking the spike/ACE2 interaction (Kadam and Wilson, 2017). Several clinical trials (IRCT20180725040596N2, NCT04260594) have demonstrated that arbidol has some effect in reducing the duration of hospitalization (Nojomi et al., 2020) and mortality of COVID-19 patients (Wang et al., 2020g). Arbidol monotherapy significantly inhibited SARS-CoV-2 viral load compared with lopinavir/ritonavir (Zhu Z. et al., 2020). Arbidol combined with LPV/r is superior to LPV/r alone (Chen C. et al., 2020). Compared with dual combination antiviral tests, the triple combination antiviral therapy of arbidol, lopinavir/litonavir and rIFNα-2b showed better therapeutic efficacy (Wei et al., 2020). However, in another clinical trial, combination with arbidol and favipiravir did not change the clinical recovery rate (Chen C. et al., 2020).
Nucleoside/nucleotide analogs (NAs), which belongs to adenine or guanine derivatives, block viral RNA synthesis by targeting viral RdRp and in a broad spectrum of RNA viruses, including human coronaviruses, human immunodeficiency virus (HIV), hepatitis B virus (HBV) and hepatitis C virus (HCV). Up to now, NAs favipiravir, ribavirin, remdesivir, galidesivir, sofosbuvir, tenofovir, NHC (β-DN4-hydroxycytidine, EIDD-1931) and EIDD-2801 have potential to treat SARS-CoV-2 (Elfiky, 2020; Sheahan et al., 2020; Wahl et al., 2021), since they tightly bind to RdRp of SARS-CoV-2 by molecular docking analysis (Elfiky, 2020). Favipiravir, an approved pyrazinecarboxamide derivative against influenza virus, can selectively and effectively inhibit the RdRp activity of RNA viruses such as influenza virus, Ebola virus, yellow fever virus, chikungunya virus, norovirus and enterovirus (Furuta et al., 2017; de Clercq, 2019). It could also block SARS CoV-2 in vitro (Wang et al., 2020c). Patients infected with SARS-CoV-2 were recruited in randomized trials to evaluate the efficacy and safety of favipiravir alone (Doi et al., 2020; Udwadia et al., 2021) or in combination with other drugs (ChiCTR2000029544) (Tu et al., 2020). Several of these trials have identified modest effect of favipiravir in shortening the time to recovery of COVID-19 patients.
Ribavirin, a guanine derivative approved in combiantion with other anti-medication for treating HCV and respiratory syncytial virus (RSV), has been evaluated in patients with SARS and MERS, but some patients may manifest side effects such as severe anemia at high doses (Zumla et al., 2016) and we do not know whether it sufficiently blocks SARS-CoV-2 (Eslami et al., 2020).
Remdesivir is a phosphoramidate prodrug of an adenine derivative and a broad-spectrum antiviral medication against pathogenic animal and human coronaviruses: bat CoVs, SARS-CoV, and MERS-CoV infection in vitro and in vivo (Sheahan et al., 2017; Warren et al., 2016). Its chemical structure is similar to that of tenofovir alafenamide, an approved HIV reverse transcriptase inhibitor. Remdesivir has been tested in a clinical for Ebola virus infected patients but showed no beneficial effect for reducing death rates compared with the control groups treated with different antibody therapies (Mulangu et al., 2019). The mechanism of action of redemsivir is that it is incorporated into nascent viral RNA chains, which results in pre-mature termination of the RNA replication by the RdRp of RNA viruses (Warren et al., 2016). Remdesivir demonstrated a potent anti-SARS-CoV-2 activity with high selectivity index values (Wang et al., 2020c). There are several successful cases of remdesivir in treating COVID-19, for example, the reported patients with mild to moderate COVID-19 were given a medication of intravenous remdesivir, and their clinical symptoms had been recovered (Grein et al., 2020). A US patient with SARS-CoV-2 recovered after receiving intravenous remdesivir in January 2020 (Holshue et al., 2020). Rhesus macaques were challenged with SARS-CoV-2 and treated with remdesivir, unlike the placebo group, macaques treated with remdesivir did not represent the severe disease observed in some patients with COVID-19 (Williamson et al., 2020). Several phase III trials were initiated in early February 2020 to evaluate the efficacy and safety of intravenous remdesivir in patients with SARS-CoV-2 (NCT04252664, NCT04257656, ISRCTN83971151, NCT04315948) (Goldman et al., 2020; Wang et al., 2020f). However, results from these trials produced mixed results which include remdesevir partially improved the symptom of wild to moderate patients or there are not significant difference in treated and untreated patients. Remdesivir, in combination of the Janus kinase inhibitor baricitinib (Kalil et al., 2021), has been granted emergency use authorization for COVID-19 by US Food and Drug Administration (FDA).
Galidesivir, an approved adenosine analog against HCV, is subsequently developed as broad-spectrum antiviral agent against yellow fever virus, Ebola virus, Zika virus, SARS and MERS-CoVs (Zumla et al., 2016). Structure-based modeling suggests that galidesivir binds to the non-catalytic site of SRAS-CoV-2 RdRp, therefore it might exert an allosteric inhibition on RdRp (Mishra and Rathore, 2021).
EIDD-2801, a prodrug of a broad-spectrum anti-CoV inhibitor NHC, a ribonucleoside analog, blocked SARS-CoV-2 infection in vitro and in vivo (Sheahan et al., 2020; Wahl et al., 2021). It also promoted pulmonary function and reduced virus titer and body weight loss of mice infected with SARS-CoV and MERS-CoV (Sheahan et al., 2020). Particularly, in human lung-only mice (LoM), which mimic the condition of human lung in a physiological context, treatment or prophylaxis administration (only one dose) of EIDD-2801 potently inhibited SARS-CoV-2 infection and pathogenesis (Wahl et al., 2021). Currently, prophylactic administration of EIDD-2801 is being tested in phase II/III clinical trial for treating COVID-19 (NCT04405570 and NCT04405739) (Cox et al., 2021).
Several approved viral protease inhibitors including disulfiram, lopinavir, and ritonavir and darunavir, are widely used to treat HIV-1 and HCV infection by selectively inhibiting viral proteases and their cleavage. Thus they have potential to inhibit coronavirus infection. Disulfiram, an alcoholism averting drug with low adverse effects, inhibited the PLpro of MERS and SARS in vitro (Lin et al., 2018). It has been reported as a non-specific Mpro inhibitor (Ma et al., 2020). But the clinical efficacy of disulfiram remains to be demonstrated. HIV protease inhibitors lopinavir and ritonavir have been shown to have no benefit for hospitalized adult patients with severe COVID-19 in a trial (Cao B. et al., 2020). Lopinavir and ritonavir improved clinical symptoms of patients with SARS in a non-randomized open-label trial (Zumla et al., 2016). HIV protease belongs to the aspartic protease family, whereas the two coronavirus proteases are from the cysteine protease family. To date, we do not know whether HIV protease inhibitors could effectively inhibit the 3CLpro and PLpro of SARS CoV-2. Furthermore, HIV protease inhibitors specifically fit the C2 symmetric pocket in the catalytic site of the HIV protease dimer, but this pocket is absent in coronavirus proteases. Darunavir, a second-generation anti-HIV-1 protease inhibitor, can inhibit SARS-CoV-2 replication in vitro, but darunavir plus cobicistat did not promote viral clearance compared with IFNα 1b treatment alone in treating COVID-19 (NCT04252274) (Chen J. et al., 2020). Darunavir or lopinavir, in combination with hydroxychloroquine, caused electrocardiogram abnormality in patients with history of cardiovascular diseases (Meriglier et al., 2021). It indicated that this combination is not safe for this group of subjects. Furthermore, in a WHO SOLIDARITY trial, remdesivir, hydroxychloroquine, lopinavir and interferon regimens appeared to have little or no effect on hospitalized COVID-19 (WHO Solidarity Trial Consortium et al., 2021).
Currently, multiple small molecules-based on structures have been identified with promisingly inhibitory effects against SARS-CoV-2. Except for RdRp as an ideal antiviral target, the main protease (Mpro) is also reported as an attractive target for inhibiting viral replication and transcription. Currently known Mpro inhibitor-based structures include N3, 11a, 11b, Camostat Mesylate, Carmofur. The crystal structure of inhibitors-Mpro complex has been reported (Wang et al., 2020d). N3, which is identified by computer-aided drug design, is a potent irreversible inhibitor of main protease and inserts into the substrate-binding pocket of Mpro according to structural analysis (Jin et al., 2020b). Dai et al. (2020) designed and synthesized two lead compounds 11a and 11b, which inhibited SARS-CoV-2 infection with low toxicity in vitro and in vivo, and with excellent pharmacokinetic properties covalently binding to the catalytic site of SARS-CoV-2 Mpro (Dai et al., 2020). In addition, combination of multiple techniques identified six Mpro inhibitors including Ebselen, Disulfiram, Tideglusib, Carmofur, Shikonin, PX-12, which inhibited enzymatic activity of PLpro and Mpro of SARS-CoV-2 and presented non-specifical anti-SARS-CoV-2 activity in vitro. The catalytic cysteine 145 of Mpro was covalently bound to 11a or 11b (Dai et al., 2020) or carmofur (Jin et al., 2020c).
Host-Targeting Antivirals
Small-molecule agents approved for other human diseases may modulate the virus–host interactions. Currently there are very few host-targeting small molecule drugs approved for antiviral purpose, with the HIV entry inhibitor maraviroc as a notable example (de Clercq and Li, 2016). Given the exploding information on the virus-host interaction, it is expected that host-targeting small molecules are the next frontier of antiviral drug discovery. Obvious advantages of these host-targeting drugs are broad-spectrum and less likelihood of drug resistance.
SARS-CoV-2 entering into host cells is key step of its life cycle, so blocking this step is critical for prevention of virus infection. Angiotensin-converting enzyme 2 (ACE2), which is highly expressed in lung, small intestine, brain, testis, kidney (Verdecchia et al., 2020), is the functional receptor of NL63 (Milewska et al., 2018), SARS-CoV (Li et al., 2003) and SARS-CoV-2 (Hoffmann et al., 2020a; Wang et al., 2020e) to facilitate their entry. ACE2 from human, monkey, pig, civet cells promotes cellular entry of SARS-CoV-2 when overexpressed, the result indicates that it is a common receptor for SARS-CoV-2 infection in these hosts (Zhou et al., 2020a). The RBD in SARS-CoV-2 spike directly binds with ACE2, thus the interaction of spike/ACE2 can be interrupted by neutralizing antibodies and small molecules. Several natural compounds, including baicalin, scutellarin, nicotianamine, and glycyrrhizin have potential to block attachment and entry of SARS-CoV-2 (Chen and Du, 2020). Glycyrrhizin, nobiletin, and neohesperidin that bind to ACE2 can partially block the binding of ACE2 and RBD (Zhou and Huang, 2020). The entry inhibitors in clinical trials have been reviewed (Oroojalian et al., 2020), which includes SSAA09E2 that blocks the interaction of ACE2-RBD. Different viruses may use the same cellular endocytic pathways to target viral entry at the point endocytosis. This strategy is promising for developing broad-spectrum antiviral drugs. There are a variety of approved drugs may have ability to block SARS-CoV-2 endocytosis in vitro (Glebov, 2020), including chlorpromazine, fluvoxamine, sertraline, promethazine, nystatin, amiloride, vinblastine, itraconazole, flubendazole, terfenadine, imipramine, beta-methyl cyclodextrin. Among them, chloroquine is a potential broad-spectrum antiviral drug against multiple virus infections (Savarino et al., 2006a; Yan et al., 2013) and widely used as anti-malarial, anti-cancer inhibitor (Savarino et al., 2006b), as well as an approved immune modulator for autoimmune diseases. Chloroquine is used for the treatment of COVID-19 as it inhibits the spread of SARS-CoV-2 in vitro (Ferner and Aronson, 2020; Touret and de Lamballerie, 2020; Wang et al., 2020c) likely by blocking endosomal acidification required for virus/cell fusion. However, chloroquine did not inhibit SARS-CoV-2 infection in human lung adenocarcinoma cell line Calu-3 overexpressing TMPRSS2 (Hoffmann et al., 2020b). An open-label trial (ChiCTR2000029609), a randomized clinical trial (NCT04323527) (Borba et al., 2020) and WHO SOLIDARITY trial were carried out to evaluate the safety and efficacy of chloroquine diphosphate against COVID-19 caused by SARS-CoV-2. The results suggest that chloroquine have little or no effect on hospitalized patients with COVID-19 (Ektorp, 2020).
Two mucosa-specific transmembrane serine protease 2 (TMPRSS2) and 4 (TMPRSS4) catalyze the cleavage of SARS-CoV-2 spike and trigger virus entry into host cells (Zang et al., 2020). Particularly, TMPRSS2 facilitates the spread and pathogenesis of SARS-CoV-2, providing a potential target for antiviral intervention. A clinical TMPRSS2 inhibitor Camostat Mesylate, potently inhibited SARS-CoV and MERS-CoV, also excellently blocked SARS-CoV-2 entry in lung adenocarcinoma cell line Calu-3 at concentrations without obvious toxicity (Hoffmann et al., 2020a). Nafamostat, a serine proteinase inhibitor, potently inhibits MERS-CoV membrane fusion relying on the activity of TMPRSS2 (Yamamoto et al., 2016) and SARS-CoV-2 infection (Wang et al., 2020c).
Ivermectin, an FDA-approved broad spectrum anti-parasitic agent (Gonzalez Canga et al., 2008),is a broad-spectrum antiviral agent with activities against flaviviruses, influenza virus and HIV-1 (Mastrangelo et al., 2012; Wagstaff et al., 2012; Lundberg et al., 2013; Tay et al., 2013; Gotz et al., 2016; Ketkar et al., 2019). It also demonstrated inhibitory effect on SARS CoV-2 infection in vitro (Caly et al., 2020). Ivermectin administration significantly reduced the mortality of COVID-19 patients (Rajter et al., 2021). Nitazoxanide, a commercial antiprotozoal agent with an antiviral potential against a broad range of viruses including human and animal coronaviruses, can also block SARS-CoV-2 replication at low micromolar concentration in vitro (Wang et al., 2020c). Although the mechanisms of actions of ivermectin and nitazoxanide are still unclear, their broad-spectrum activities suggest host-targeting mechanisms. Further in vivo and clinical evaluations of these drugs are warranted.
Pegylated interferon alfa-2a and - 2b alone and in combination with other antiviral agents, approved for treating HBV and HCV infection, could be used to stimulate innate antiviral responses in COVID-19 patients. One research showed that type I and type III IFN have potential inhibitory effects on SARS-CoV-2 in vitro (Felgenhauer et al., 2020). However, several clinical trials involving interferons showed that interferon regimens have little or no effect on hospitalized COVID-19 patients (ChiCTR2000029387, ISRCTN83971151, NCT04315948) (Zeng Y.M. et al., 2020; WHO Solidarity Trial Consortium et al., 2021).
Host RNA binding proteins are emerging host factors that are involved in the life cycle of viruses. Recently, Sun et al. reported in vivo genomic RNA structure landscape of SARS-CoV-2, and validated several key structure elements influencing viral protein translation and abundance of virus sub-genomic structure. Using deep learning algorithm based on the information of the viral genomic structure, they predicted 42 host binding proteins on SARS-CoV-2 RNA, such as DDX24, NPM1. Further, they identified antisense oligonucleotides (ASOs) targeting viral RNA structure units and FDA-approved drugs Deguelin, Nilotinib, and Sorafenib inhibiting the predicted RNA binding protein expression and showed these compounds decreased SARS-CoV-2 infection in human cells (Sun et al., 2021).
Traditional Chinese Medicine
Traditional Chinese medicine including plant extracts present potential anti-coronavirus ability for therapeutic development (Li et al., 2005; Park et al., 2017). The active components from traditional Chinese herbs can treat COVID-19 by controlling virus infection and regulating immune response, inflammatory reaction and hypoxia response (Zhang et al., 2020). Combination of Chinese herbs (Lianhuaqingwen capsule) and western medicines have been encouraged to treat infected patients in hospital with good efficacy in China (Xia et al., 2020c). One review summarized natural products in the role of anti-SARS-CoV-2 infection (Zhou and Huang, 2020), including viral targets, natural components and mechanism of action against SARS-CoV-2.
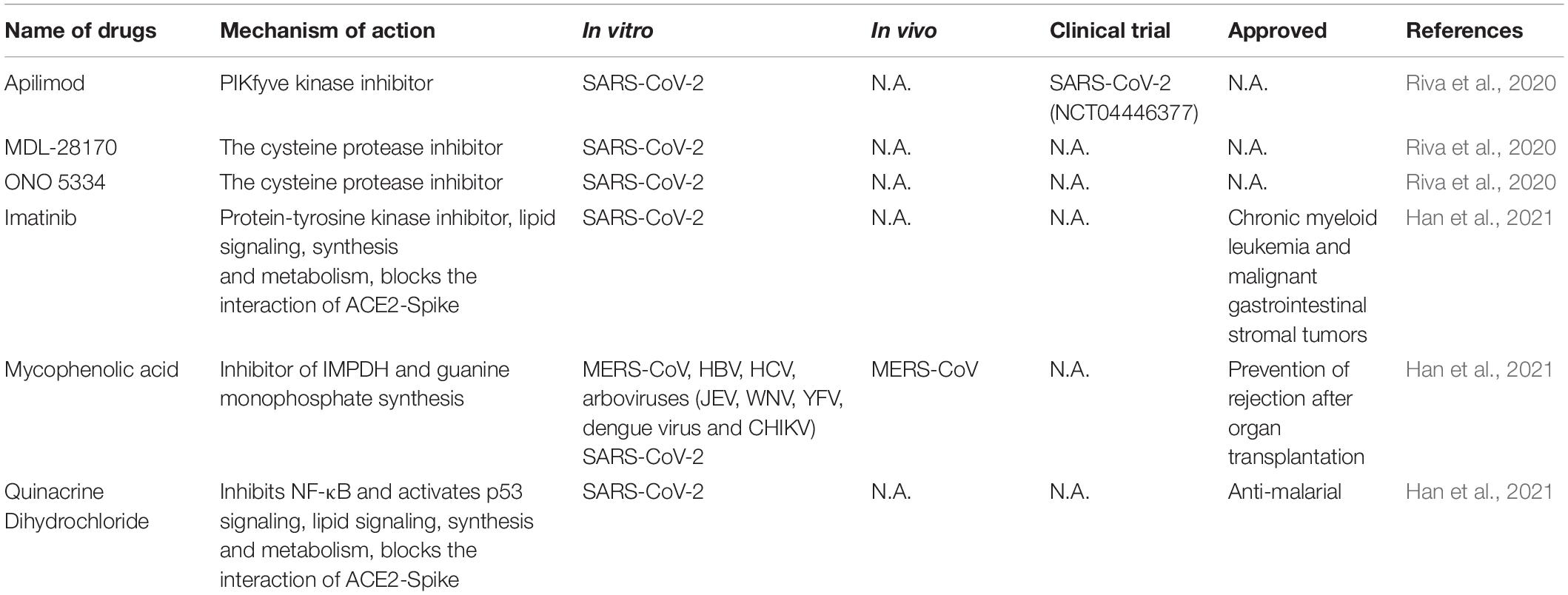
High-Throughput Screening for Anti-SARS-CoV-2 Drugs
Large scale repurposing screening of known or approved drugs could significantly accelerate the deployment of novel therapies for COVID-19 (Table 3). One of such efforts is profiling FDA-approved small molecules LOPAC-1280 and ReFRAME (Repurposing, Focused Rescue, and Accelerated Medchem) drug library containing about 12,000 bioactive molecules (Riva et al., 2020). A total of 21 known drugs showed inhibition of SARS CoV-2 replication in dose-dependent manners. All of the PIKfyve kinase inhibitor apilimod, the cysteine protease inhibitors MDL-28170 and ONO 5334 presented an efficacious potency against SARS-CoV-2 replication in iPSC cells and apilimod efficiently inhibited SARS-CoV-2 infection in human lung tissues.
In another screening effort, lung and colonic organoid model derived from human pluripotent stem cells were used as the infection target, presumably more physiologically relevant than using cancer cell lines. From this screening, imatinib, mycophenolic acid and quinacrine dihydrochloride were identified from FDA-approved library as SARS-CoV-2 entry inhibitors at physiologically relevant concentrations. Imatinib and QNHC bind with ACE2 with a high affinity based on surface plasmon resonance binding analysis. Imatinib is related to lipid signaling, synthesis and metabolism based on RNA-Seq analysis, therefore likely affects SARS-CoV-2 by modulating host responses (Han et al., 2021).
Artificial Intelligence-Based Drug Discovery for Anti-SARS-CoV-2
In the big data era, artificial intelligence (AI) algorithms are increasingly being applied for rapid and cost-effective drug discovery (Fleming, 2018). The scale and efficiency that AI brings to drug discovery is especially relevant for treating COVID-19 epidemic. Thus, it supplies a good opportunity for drug discovery and development via introducing AI tools and network medicine, for example, drug target identification based on protein-protein interactome (Zhou et al., 2020d). In one such studies, 332 interactions have been identified between SARS-CoV-2 proteins and human host proteins including ACE2, Furin, TEPRSS2, NRP1, eEF1A, etc. (Gordon et al., 2020). Among of the identified host proteins, 69 druggable targets were selected and two inhibitors can potently block SARS-CoV-2 infection, one by inhibiting mRNA translation (Zotatifin) and the other by regulating sigma-2 and sigma-2 receptors (haloperidol). Also derived from this study, the inhibitor plitidepsin targeting host protein eEFIA, which interacts with multiple coronavirus proteins, potently inhibits SARS-CoV-2 infection in vivo (White et al., 2021). From another study, 16 potential anti-coronavirus drugs (Irbesartan, toremifene, camphor, equilin, mesalazine, mercaptopurine, paroxetine, sirolimus, carvedilol, colchicine, dactinomycin, melatonin, quinacrine, eplerenone, emodin, Oxymetholone) were found to inhibit SARS-CoV-2 infection based on drug targets and viral-host interactions through implementing a systemic network medicine platform (Zhou et al., 2020c). Among of them, toremifene, an approved selective estrogenic receptor modulator for treating breast cancer, can inhibit various viral infection including MERS-CoV (Cong et al., 2018), SARS-CoV (Dyall et al., 2014), and SARS-CoV-2 (Jeon et al., 2020) by blocking the interaction of ACE2-Spike and Nsp14 of SARS-CoV-2 based on computational biophysics analysis (Martin and Cheng, 2020). Beck et al. used deep learning-based drug target prediction algorithm to predict that several commercially available drugs atazanavir, remdesivir, efavirenz, ritonavir, and dolutegravir potently inhibits the activity of 3CLpro of SARS-CoV-2 with low Kd values (Beck et al., 2020). Gysi et al. (2020) identified 81 potential repurposing compounds against SARS-CoV-2 via graph neural network model. Zeng X. et al. (2020) used graph representation learning techniques to identify 41 potential candidates against SARS-CoV-2 infection including niclosamide, melatonin and dexamethasone.
Stebbing et al. used AI-algorithms to identify baricitinib, an approved Janus kinase (JAK)1/JAK2 inhibitor for treating rheumatoid arthritis, to be effective at inhibiting SARS-CoV-2 infection as well as reducing virus induced inflammations (Stebbing et al., 2020). Although baricitinib can cause potential adverse effect including lymphopenia, anemia and increase co-infection crisis with other pathogens (Pujari et al., 2020), its combination with remdesivir is superior to remdesivir alone in improving the clinical recovery rate of patients with COVID-19 with few serious side effects (Kalil et al., 2021). Ge Y. et al. (2021) used machine learning and statistical analysis to uncover a ploly-ADP-ribose polymerase (PARP1) inhibitor meguparib (CVL218) which reduced SARS-CoV-2 replication without toxic effects in vitro. In addition it also suppressed IL-6 production. Molecular docking simulation showed that meguparib can bind to the nucleoprotein of SARS-CoV-2, which might mediate the inhibition of viral replication (Ge Y. et al., 2021).
In silico molecular modeling suggests that several FAD-approved anticancer drugs (Capmatinib, Pemigatinib, Selpercatinib, and Tucatinib) might be able to inhibit COVID-19 by docking on Mpro and spike of SARS-CoV-2 (Parveen and Alnoman, 2021). Network medicine methods suggest potential drug combinations (anti-inflammatory plus anti-viral drug) for treating COVID-19, including toremifene plus emodin, mercaptopurine plus melatonin, and sirolimus plus dactinomycin (Zhou et al., 2020c). Another network medicine analysis predicted that anti-viral inhibitor toremifene plus anti-inflammatory drug melatonin have potential to treat COVID-19 (Cheng et al., 2020).
Above mentioned AI-based drugs against COVID-19 are encouraging, effective and robust experimental evaluations in vitro and in vivo will further increase the successful rate for drug discovery in preclinical and clinical trials. AI-tools also integrate pharmocogenetic and pharmocogenomic information to figure out the key genetic targets and therapies against COVID-19.
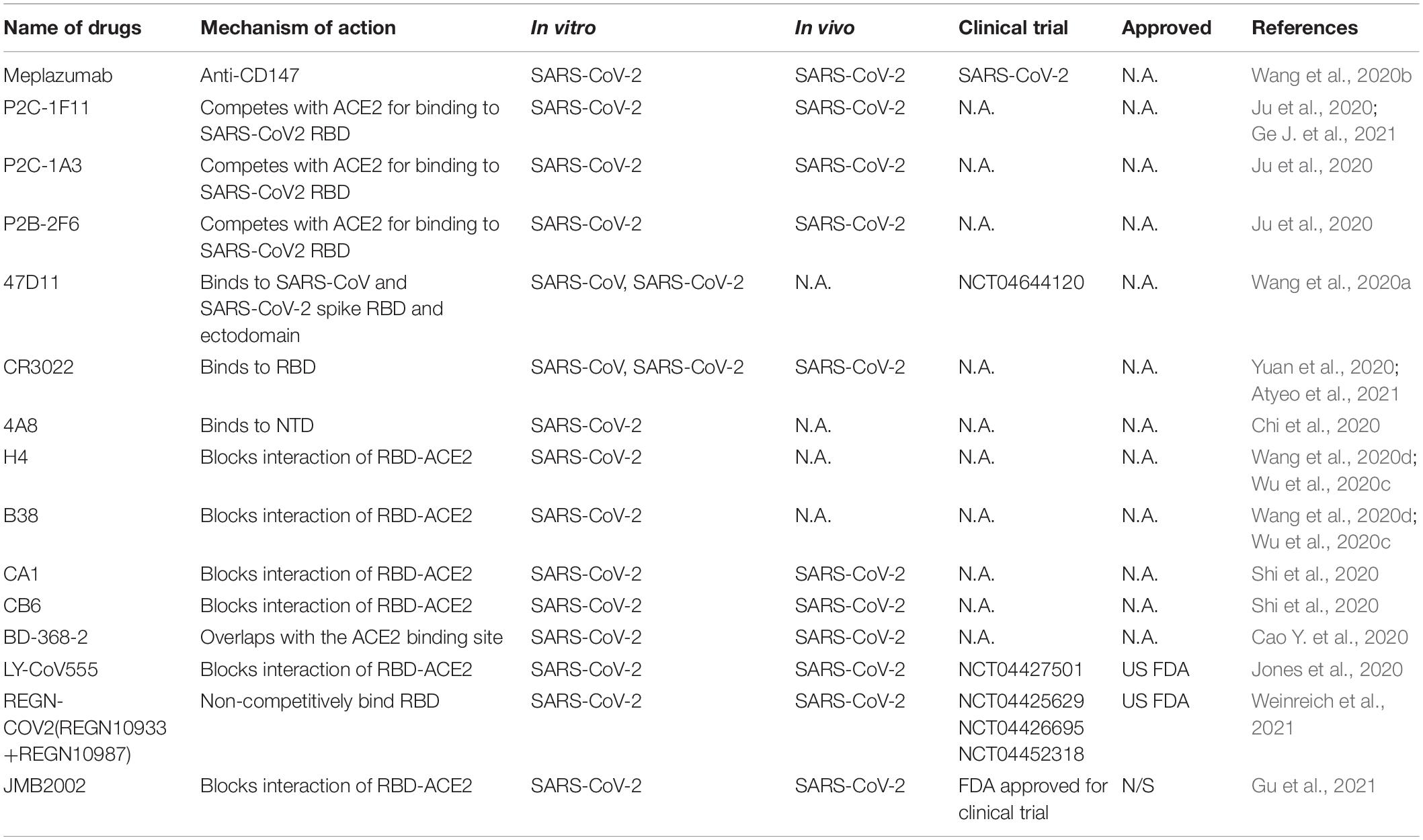
Therapeutic Antibodies Against SARS-CoV-2
Responsible for binding the receptor and mediating cellular entry, the spike protein of SARS CoVs is the ideal target for neutralizing antibodies as therapy (Table 4). Several isolated monoclonal antibodies from SARS-CoV-2 infected patients in recovery and convalescent periods can recognize receptor-binding domain (RBD), N terminal domain (NTD) and S2 domain of spike. In one such efforts, Ju et al. (2020) isolated three potent neutralizing antibodies P2C-1F11, P2C-1A3, and P2B-2F6. The three antibodies compete with ACE2 for binding to SARS-CoV2 RBD without cross-reacting with plasma from SARS-CoV and MERS-CoV patients. Among the three antibodies, P2C-1F11 displays the most potent neutralizing activity in vitro and in vivo. It occupies a large binding surface on RBD and triggers quick and extensive shedding of spike protein from cell surface, thus neutralizes the virus (Ge J. et al., 2021). From humanized mouse model, a monoclonal antibody 47D11 was identified to bind to SARS-CoV and SARS-CoV-2 spike RBD and ectodomain with similar affinities, and the binding was not competed by ACE2 (Wang et al., 2020a). However, three reported SARS-CoV RBD-targeting mABs S230, m396, and 80R have very low cross-reactivity at different concentrations compared with SARS-CoV-2 for different antigenicity (Wrapp et al., 2020). Another reported SARS-CoV-specific antibody CR3022 derived from convalescent SARS-CoV infected patient also potently binds to RBD of SARS-CoV-2. CR3022 binds SARS-CoV-2 spike at a site that is different from the ACE2 binding site. Combination of CR3022 and other antibodies which has their epitopes in RBD, can synergistically neutralize SARS-CoV-2 (Yuan et al., 2020). Chi et al. isolated a monoclonal antibody 4A8 from convalescent patient that potently neutralizes both authentic SARS-CoV-2 and SARS-CoV-2 pseudovirus. They utilized Cryo–electron microscopy, and obtained a structure with a resolution of 3.1 angstroms for the epitope 4A8 - NTD interface (Chi et al., 2020). These results indicate that combination of RBD and NTD-targeting antibodies may be useful as therapeutic cocktails.
Additional potent monoclonal antibodies for specific RBD or S1 subunits were also identified in vitro and in vivo and their structures with spike or ACE2 have been deciphered (Wang et al., 2020d), these antibody includes H4, B38 (Wu et al., 2020c), CA1, CB6 (Shi et al., 2020), BD-368-2 (Cao Y. et al., 2020), and cocktail of REGN10987 and REGN10933 (Hansen et al., 2020). Still some antibodies can bind to the spike to reduce SARS-CoVs titer, but is unable to block the binding of virus and ACE2, such as n3130, n3088 (Wu et al., 2020b), and S309 from memory B cells of recovered from a patient infected with SARS-CoV, indicating multiple mechanisms of neutralizations (Pinto et al., 2020).
Clinical trials with neutralizing antibodies have been conducted extensively. In a phase II clinical trial (NCT04427501), 2,800 mg dose of SARS-CoV-2 neutralizing antibody LY-CoV555 from Lilly declined the viral load in patients (Chen et al., 2021), but LY-CoV555 in combination of remdesivir have no efficacy in hospitalized patients with COVID-19(NCT04501978) (ACTIV-3/TICO LY-CoV555 Study Group et al., 2021). A neutralizing antibody cocktail REGN-COV2 reduced viral load and enhanced immune responses (NCT04425629) (Weinreich et al., 2021). Due to the positive responses, both antibodies were authorized by US FDA for emergency use. A human anti-SARS-CoV-2 RBD antibody JMB2002, can prevent and treat SARS-CoV-2 infection in rhesus macaque by blocking the binding of ACE2 to RBD of multiple variants including the South African mutant (B.1.351), the UK mutant (B.1.1.7) and the Brazilian mutant (P.1) (Gu et al., 2021). Recently, JMB2002 was approved by the US FDA for clinical trial. To date, dozens of antibodies targeting SARS-CoV-2 spike are still undergoing clinical trials for COVID-19 treatment, and many more antibodies are in discovery research (DeFrancesco, 2020; Yang et al., 2020).
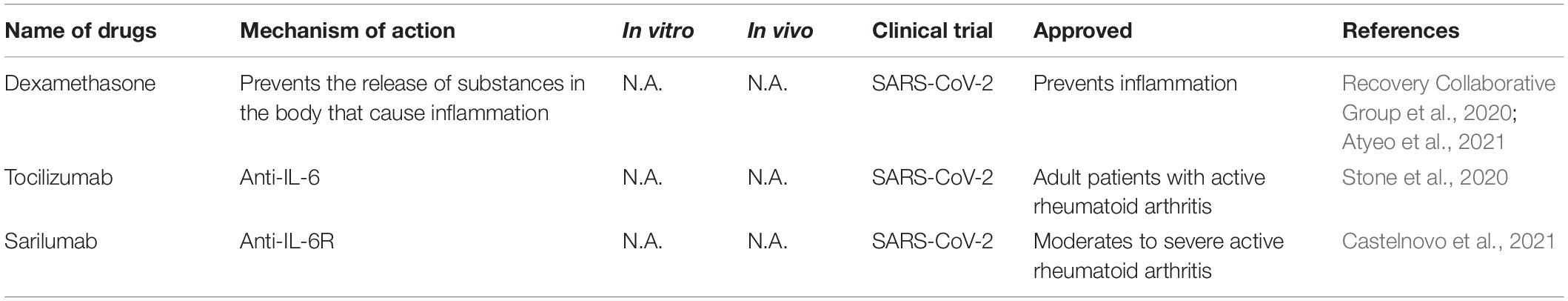
Immuno-Regulators Against SARS-CoV-2
SARS-CoV-2 infection causes severe acute pneumonic processes with pathological damages, including inflammatory cytokine storm characterized by heightened levels of C-reactive protein, ferritin, interleukin (IL)-1, IL-6, IL-8, and TNF-α (del Valle et al., 2020; Huang et al., 2020; Moore and June, 2020; Ruan et al., 2020). Several immune interventions mitigated host organ failure in the viral pneumonia (Table 5), such as the use of corticosteroid, but there is still some debate in these uses (Russell et al., 2020; Shang et al., 2020; Zhou et al., 2020b). Supportive use of corticosteroid produced a positive effect on COVID-19 with lower mortality before the development of acute respiratory distress syndrome (ARDS) (Boglione et al., 2021). Another large-scale randomized clinical trials (NCT04381936) reported that the use of glucocorticoid dexamethasone reduced 28-day mortality of hospitalized COVID-19 patients (Recovery Collaborative Group et al., 2020). In hospitalized COVID-19 patients with symptoms of pneumonitis or hypoxia, dexamethasone or remdesivir plus baricitini have shown benefits in randomized controlled trials (Calabrese and Calabrese, 2021).
Dampening IL6 is a potential therapeutic avenue for immuno-modulation because IL6 is elevated in patients suffering from ARDS and its level correlated positively with viral loads (del Valle et al., 2020). Tocilizumab, a monoclonal anti-IL-6 antibody, is a promising anti-inflammatory regent in the treatment of COVID-19 but the results are mixed results from clinical trials. A randomized, double-blind, placebo-controlled trial (NCT04356937) show that tocilizumab has no efficacy for improving statement of treatment in moderately hospitalized patients infected with SARS-CoV-2 (Stone et al., 2020). Tocilizumab and sarilumab, an IL-6R humanized monoclonal antibody, have been evaluated in clinical trials (NCT04330638, NCT04486521, NCT04329650) by decreasing IL6 level to decrease the risk of mortality caused by COVID-19 (Castelnovo et al., 2021).
Discussion
In order to cope with emerging and re-emerging infectious diseases, challenges and opportunities abound for developing antiviral therapeutics. A large number of anti-viral agents are currently being explored for treating SARS-CoV-2 infection. Repurposing of existing drugs have demonstrated power by bringing several drugs to approval for treating COVID-19 patients, such as remdesivir and dexamethasone. However, these drugs still suffer from suboptimal therapeutic effect or known strong side-effect. Direct acting antivirals (DAAs) are rapidly being developed and there is great hope for producing highly potent antivirals from these efforts. Structure-based studies have significantly boosted the efficiency and precision of these developments. However, drug resistant strains are going to arise sooner or later. The recent rises of several high transmissible strains sounded alarms for currently used vaccines and neutralizing antibodies. Combinations of DAAs or neutralizing antibodies will likely be required for effective control of the viruses, much like the development of HIV cocktail therapies. On the other hand, development of broad-spectrum antiviral drugs not only fight against COVID-19 but also provide arsenals for protection from future viral outbreaks. The wealth of knowledge that has been and continues to be gleaned on the interaction between viruses and hosts will guide and prod the development of host-targeting antivirals, which have the potential advantages of broad-spectrum therapeutic effect and insensitivity to viral evasion. Antiviral drug development in general will benefit from the heightened level of social awareness of the COVID-19 epidemic and hopefully provide a safeguard against future viral epidemics for the humankind.
Author Contributions
MM and XT wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by Spring Breeze Fund of Tsinghua University, the China National Funds for Excellent Young Scientists (31722030) to XT and grants from the Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure to XT. MM is supported by a postdoctoral fellowship from Tsinghua-Peking Center for Life Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
ACTIV-3/TICO LY-CoV555 Study Group, Lundgren, J. D., Grund, B., Barkauskas, C. E., Holland, T. L., Gottlieb, R. L., et al. (2021). A neutralizing monoclonal antibody for hospitalized patients with Covid-19. N. Engl. J. Med. 384, 905–914. doi: 10.1056/nejmoa2033130
Atyeo, C., Slein, M. D., Fischinger, S., Burke, J., Schafer, A., Leist, S. R., et al. (2021). Dissecting strategies to tune the therapeutic potential of SARS-CoV-2-specific monoclonal antibody CR3022. JCI Insight 6:e143129.
Beck, B. R., Shin, B., Choi, Y., Park, S., and Kang, K. (2020). Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 18, 784–790. doi: 10.1016/j.csbj.2020.03.025
Blaising, J., Polyak, S. J., and Pecheur, E. I. (2014). Arbidol as a broad-spectrum antiviral: an update. Antiviral Res. 107, 84–94. doi: 10.1016/j.antiviral.2014.04.006
Boglione, L., Rostagno, R., Poletti, F., Moglia, R., Bianchi, B., Esposito, M., et al. (2021). The proper use of corticosteroids for 2019-nCov pneumonia: towards promising results? J. Infect. 82, e6–e7.
Borba, M. G. S., Val, F. F. A., Sampaio, V. S., Alexandre, M. A. A., Melo, G. C., Brito, M., et al. (2020). Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw. Open 3:e208857.
Breining, P., Frolund, A. L., Hojen, J. F., Gunst, J. D., Staerke, N. B., Saedder, E., et al. (2021). Camostat mesylate against SARS-CoV-2 and COVID-19-rationale, dosing and safety. Basic Clin. Pharmacol. Toxicol. 128, 204–212. doi: 10.1111/bcpt.13533
Calabrese, L. H., and Calabrese, C. (2021). Baricitinib and dexamethasone for hospitalized patients with COVID-19. Clevel. Clin. J. Med. 1–3. doi: 10.3949/ccjm.88a.ccc073
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., and Wagstaff, K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 178:104787. doi: 10.1016/j.antiviral.2020.104787
Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., et al. (2020). A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 382, 1787–1799.
Cao, Y., Su, B., Guo, X., Sun, W., Deng, Y., Bao, L., et al. (2020). Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B Cells. Cell 182, 73–84.e16.
Castelnovo, L., Tamburello, A., Lurati, A., Zaccara, E., Marrazza, M. G., Olivetti, M., et al. (2021). Anti-IL6 treatment of serious COVID-19 disease: a monocentric retrospective experience. Medicine 100:e23582. doi: 10.1097/md.0000000000023582
Chen, C., Zhang, Y., Huang, J., Yin, P., Cheng, Z., Wu, J., et al. (2020). Favipiravir versus Arbidol for COVID-19: a randomized clinical trial. medRxiv [Preprint]. doi: 10.1101/2020031720037432
Chen, H., and Du, Q. (2020). Potential natural compounds for preventing SARS-CoV-2 (2019-nCoV) infection. Preprints.
Chen, J., Xia, L., Liu, L., Xu, Q., Ling, Y., Huang, D., et al. (2020). Antiviral activity and safety of darunavir/cobicistat for the treatment of COVID-19. Open Forum Infect. Dis. 7:ofaa241.
Chen, P., Nirula, A., Heller, B., Gottlieb, R. L., Boscia, J., Morris, J., et al. (2021). SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 384, 229–237. doi: 10.1056/nejmoa2029849
Cheng, F., Rao, S., and Mehra, R. (2020). COVID-19 treatment: combining anti-inflammatory and antiviral therapeutics using a network-based approach. Cleve. Clin. J. Med. 1–6. doi: 10.3949/ccjm.87a.ccc037
Chi, X., Yan, R., Zhang, J., Zhang, G., Zhang, Y., Hao, M., et al. (2020). A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science 369, 650–655.
Cong, Y., Hart, B. J., Gross, R., Zhou, H., Frieman, M., Bollinger, L., et al. (2018). MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS One 13:e0194868. doi: 10.1371/journal.pone.0194868
Cox, R. M., Wolf, J. D., and Plemper, R. K. (2021). Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 6, 11–18. doi: 10.1038/s41564-020-00835-2
Dai, W., Zhang, B., Jiang, X. M., Su, H., Li, J., Zhao, Y., et al. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368, 1331–1335.
de Clercq, E. (2019). New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chem. Asian J. 14, 3962–3968. doi: 10.1002/asia.201900841
de Clercq, E., and Li, G. (2016). Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 29, 695–747. doi: 10.1128/cmr.00102-15
DeFrancesco, L. (2020). COVID-19 antibodies on trial. Nat. Biotechnol. 38, 1242–1252. doi: 10.1038/s41587-020-0732-8
del Valle, D. M., Kim-Schulze, S., Huang, H. H., Beckmann, N. D., Nirenberg, S., Wang, B., et al. (2020). An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643.
Doi, Y., Hibino, M., Hase, R., Yamamoto, M., Kasamatsu, Y., Hirose, M., et al. (2020). A prospective, randomized, open-label trial of early versus late favipiravir therapy in hospitalized patients with COVID-19. Antimicrob. Agents Chemother. 64:e01897-20.
Dyall, J., Coleman, C. M., Hart, B. J., Venkataraman, T., Holbrook, M. R., Kindrachuk, J., et al. (2014). Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 58, 4885–4893. doi: 10.1128/aac.03036-14
Ektorp, E. (2020). Death threats after a trial on chloroquine for COVID-19. Lancet Infect. Dis. 20:661. doi: 10.1016/s1473-3099(20)30383-2
Elfiky, A. A. (2020). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 253:117592. doi: 10.1016/j.lfs.2020.117592
Eslami, G., Mousaviasl, S., Radmanesh, E., Jelvay, S., Bitaraf, S., Simmons, B., et al. (2020). The impact of sofosbuvir/daclatasvir or ribavirin in patients with severe COVID-19. J. Antimicrob. Chemother. 75, 3366–3372. doi: 10.1093/jac/dkaa331
Felgenhauer, U., Schoen, A., Gad, H. H., Hartmann, R., Schaubmar, A. R., Failing, K., et al. (2020). Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 295, 13958–13964. doi: 10.1074/jbc.ac120.013788
Ferner, R. E., and Aronson, J. K. (2020). Chloroquine and hydroxychloroquine in covid-19. BMJ 369:m1432. doi: 10.1136/bmj.m1432
Furuta, Y., Komeno, T., and Nakamura, T. (2017). Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 449–463. doi: 10.2183/pjab.93.027
Ge, J., Wang, R., Ju, B., Zhang, Q., Sun, J., Chen, P., et al. (2021). Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 12:250.
Ge, Y., Tian, T., Huang, S., Wan, F., Li, J., Li, S., et al. (2021). A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. Signal Transduct. Target. Ther. 6:165. doi: 10.1038/s41392-021-00568-6
Glebov, O. O. (2020). Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 287, 3664–3671. doi: 10.1111/febs.15369
Goldman, J. D., Lye, D. C. B., Hui, D. S., Marks, K. M., Bruno, R., Montejano, R., et al. (2020). Remdesivir for 5 or 10 days in patients with severe Covid-19. N. Engl. J. Med. 383, 1827–1837.
Gonzalez Canga, A., Sahagun Prieto, A. M., Diez Liebana, M. J., Fernandez Martinez, N., Sierra Vega, M., and Garcia Vieitez, J. J. (2008). The pharmacokinetics and interactions of ivermectin in humans–a mini-review. AAPS J. 10, 42–46. doi: 10.1208/s12248-007-9000-9
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468.
Gotz, V., Magar, L., Dornfeld, D., Giese, S., Pohlmann, A., Hoper, D., et al. (2016). Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 6:23138.
Grein, J., Ohmagari, N., Shin, D., Diaz, G., Asperges, E., Castagna, A., et al. (2020). Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 382, 2327–2336.
Gu, C., Cao, X., Wang, Z., Hu, X., Yao, Y., Zhou, Y., et al. (2021). A human antibody with blocking activity to RBD proteins of multiple SARS-CoV-2 variants including B.1.351 showed potent prophylactic and therapeutic efficacy against SARS-CoV-2 in rhesus macaques. bioRxiv [Preprint]. doi: 10.1101/2021.02.07.429299
Gysi, D. M., Do Valle, I., Zitnik, M., Ameli, A., Gan, X., Varol, O., et al. (2020). Network medicine framework for identifying drug repurposing opportunities for COVID-19. arXiv [Preprint]. arXiv:2004.07229v1
Han, Y., Duan, X., Yang, L., Nilsson-Payant, B. E., Wang, P., Duan, F., et al. (2021). Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275.
Hansen, J., Baum, A., Pascal, K. E., Russo, V., Giordano, S., Wloga, E., et al. (2020). Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014.
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Kruger, N., Herrler, T., Erichsen, S., et al. (2020a). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8.
Hoffmann, M., Mosbauer, K., Hofmann-Winkler, H., Kaul, A., Kleine-Weber, H., Kruger, N., et al. (2020b). Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 585, 588–590. doi: 10.1038/s41586-020-2575-3
Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936.
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506.
Ita, K. (2021). Coronavirus disease (COVID-19): current status and prospects for drug and vaccine development. Arch. Med. Res. 52, 15–24. doi: 10.1016/j.arcmed.2020.09.010
Jeon, S., Ko, M., Lee, J., Choi, I., Byun, S. Y., Park, S., et al. (2020). Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 64:e00819-20.
Jin, Y., Yang, H., Ji, W., Wu, W., Chen, S., Zhang, W., et al. (2020a). Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses 12:372. doi: 10.3390/v12040372
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020b). Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293.
Jin, Z., Zhao, Y., Sun, Y., Zhang, B., Wang, H., Wu, Y., et al. (2020c). Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 27, 529–532. doi: 10.1038/s41594-020-0440-6
Jones, B. E., Brown-Augsburger, P. L., Corbett, K. S., Westendorf, K., Davies, J., Cujec, T. P., et al. (2020). LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv [Preprint]. doi: 10.1101/2020.09.30.318972
Ju, B., Zhang, Q., Ge, J., Wang, R., Sun, J., Ge, X., et al. (2020). Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119.
Kadam, R. U., and Wilson, I. A. (2017). Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U.S.A. 114, 206–214. doi: 10.1073/pnas.1617020114
Kalil, A. C., Patterson, T. F., Mehta, A. K., Tomashek, K. M., Wolfe, C. R., Ghazaryan, V., et al. (2021). Baricitinib plus remdesivir for hospitalized adults with Covid-19. N. Engl. J. Med. 384, 795–807.
Ketkar, H., Yang, L., Wormser, G. P., and Wang, P. (2019). Lack of efficacy of ivermectin for prevention of a lethal Zika virus infection in a murine system. Diagn. Microbiol. Infect. Dis. 95, 38–40. doi: 10.1016/j.diagmicrobio.2019.03.012
Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., et al. (2003). A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348, 1986–1994.
Li, G., and de Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 19, 149–150. doi: 10.1038/d41573-020-00016-0
Li, S. Y., Chen, C., Zhang, H. Q., Guo, H. Y., Wang, H., Wang, L., et al. (2005). Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 67, 18–23. doi: 10.1016/j.antiviral.2005.02.007
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454.
Liao, W., Liu, X., Yang, Q., Liu, H., Liang, B., Jiang, J., et al. (2020). Deguelin inhibits HCV replication through suppressing cellular autophagy via down regulation of Beclin1 expression in human hepatoma cells. Antiviral Res. 174:104704. doi: 10.1016/j.antiviral.2020.104704
Lim, S. Y., Osuna, C. E., Best, K., Taylor, R., Chen, E., Yoon, G., et al. (2020). A direct-acting antiviral drug abrogates viremia in Zika virus-infected rhesus macaques. Sci. Transl. Med. 12:eaau9135. doi: 10.1126/scitranslmed.aau9135
Lin, M. H., Moses, D. C., Hsieh, C. H., Cheng, S. C., Chen, Y. H., Sun, C. Y., et al. (2018). Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Res. 150, 155–163. doi: 10.1016/j.antiviral.2017.12.015
Lundberg, L., Pinkham, C., Baer, A., Amaya, M., Narayanan, A., Wagstaff, K. M., et al. (2013). Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis virus replication. Antiviral Res. 100, 662–672. doi: 10.1016/j.antiviral.2013.10.004
Ma, C., Hu, Y., Townsend, J. A., Lagarias, P. I., Marty, M. T., Kolocouris, A., et al. (2020). Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Transl. Sci. 3, 1265–1277. doi: 10.1021/acsptsci.0c00130
Martin, W. R., and Cheng, F. (2020). Repurposing of FDA-approved toremifene to treat COVID-19 by blocking the spike glycoprotein and NSP14 of SARS-CoV-2. J. Proteome Res. 19, 4670–4677. doi: 10.1021/acs.jproteome.0c00397
Mastrangelo, E., Pezzullo, M., De Burghgraeve, T., Kaptein, S., Pastorino, B., Dallmeier, K., et al. (2012). Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J. Antimicrob. Chemother. 67, 1884–1894. doi: 10.1093/jac/dks147
Meriglier, E., Rivoisy, C., Hessamfar, M., Bernard, N., Aureau, I., Lapoirie, J., et al. (2021). Safety of hydroxychloroquine and darunavir or lopinavir in COVID-19 infection. J. Antimicrob. Chemother. 76, 482–486. doi: 10.1093/jac/dkaa441
Milewska, A., Nowak, P., Owczarek, K., Szczepanski, A., Zarebski, M., Hoang, A., et al. (2018). Entry of human coronavirus NL63 into the Cell. J. Virol. 92:e01933-17.
Mishra, A., and Rathore, A. S. (2021). RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2. J. Biomol. Struct. Dyn. 1–13. doi: 10.1080/07391102.2021.1875886
Moore, J. B., and June, C. H. (2020). Cytokine release syndrome in severe COVID-19. Science 368, 473–474. doi: 10.1126/science.abb8925
Morse, J. S., Lalonde, T., Xu, S., and Liu, W. R. (2020). Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem 21, 730–738. doi: 10.1002/cbic.202000047
Mulangu, S., Dodd, L. E., Davey, R. T. Jr., Tshiani Mbaya, O., Proschan, M., Mukadi, D., et al. (2019). A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 381, 2293–2303.
Nojomi, M., Yassin, Z., Keyvani, H., Makiani, M. J., Roham, M., Laali, A., et al. (2020). Effect of Arbidol (Umifenovir) on COVID-19: a randomized controlled trial. BMC Infect. Dis. 20:954. doi: 10.1186/s12879-020-05698-w
Nukui, M., O’Connor, C. M., and Murphy, E. A. (2018). The natural flavonoid compound deguelin inhibits HCMV lytic replication within fibroblasts. Viruses 10:614. doi: 10.3390/v10110614
Oany, A. R., Mia, M., Pervin, T., Junaid, M., Hosen, S. M. Z., and Moni, M. A. (2020). Design of novel viral attachment inhibitors of the spike glycoprotein (S) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) through virtual screening and dynamics. Int. J. Antimicrob. Agents 56:106177. doi: 10.1016/j.ijantimicag.2020.106177
Oroojalian, F., Haghbin, A., Baradaran, B., Hemmat, N., Shahbazi, M. A., Baghi, H. B., et al. (2020). Novel insights into the treatment of SARS-CoV-2 infection: an overview of current clinical trials. Int. J. Biol. Macromol. 165, 18–43. doi: 10.1016/j.ijbiomac.2020.09.204
Park, J. Y., Yuk, H. J., Ryu, H. W., Lim, S. H., Kim, K. S., Park, K. H., et al. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 32, 504–515. doi: 10.1080/14756366.2016.1265519
Parveen, S., and Alnoman, R. B. (2021). Potential exploration of recent FDA-approved anticancer drugs against models of SARS-CoV-2′s main protease and spike glycoprotein: a computational study. Biointerface Res. Appl. 11, 10059–10073. doi: 10.33263/briac113.1005910073
Pinto, D., Park, Y. J., Beltramello, M., Walls, A. C., Tortorici, M. A., Bianchi, S., et al. (2020). Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295.
Pujari, R., Thommana, M. V., Ruiz Mercedes, B., and Serwat, A. (2020). Therapeutic options for COVID-19: a review. Cureus 12:e10480.
Rajter, J. C., Sherman, M. S., Fatteh, N., Vogel, F., Sacks, J., and Rajter, J. J. (2021). Use of ivermectin is associated with lower mortality in hospitalized patients with coronavirus disease 2019: the ivermectin in COVID nineteen study. Chest 159, 85–92. doi: 10.1016/j.chest.2020.10.009
Recovery Collaborative Group, Horby, P., Lim, W. S., Emberson, J. R., Mafham, M., Bell, J. L., et al. (2020). Dexamethasone in hospitalized patients with Covid-19 - preliminary report. N. Engl. J. Med. 384, 693–704.
Riva, L., Yuan, S., Yin, X., Martin-Sancho, L., Matsunaga, N., Pache, L., et al. (2020). Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 586, 113–119.
Ruan, Q., Yang, K., Wang, W., Jiang, L., and Song, J. (2020). Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 46, 846–848. doi: 10.1007/s00134-020-05991-x
Russell, C. D., Millar, J. E., and Baillie, J. K. (2020). Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 395, 473–475. doi: 10.1016/s0140-6736(20)30317-2
Savarino, A., di Trani, L., Donatelli, I., Cauda, R., and Cassone, A. (2006a). New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 6, 67–69. doi: 10.1016/s1473-3099(06)70361-9
Savarino, A., Lucia, M. B., Giordano, F., and Cauda, R. (2006b). Risks and benefits of chloroquine use in anticancer strategies. Lancet Oncol. 7, 792–793. doi: 10.1016/s1470-2045(06)70875-0
Shang, L., Zhao, J., Hu, Y., Du, R., and Cao, B. (2020). On the use of corticosteroids for 2019-nCoV pneumonia. Lancet 395, 683–684. doi: 10.1016/s0140-6736(20)30361-5
Sheahan, T. P., Sims, A. C., Graham, R. L., Menachery, V. D., Gralinski, L. E., Case, J. B., et al. (2017). Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9:eaal3653. doi: 10.1126/scitranslmed.aal3653
Sheahan, T. P., Sims, A. C., Zhou, S., Graham, R. L., Pruijssers, A. J., Agostini, M. L., et al. (2020). An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12:eabb5883. doi: 10.1126/scitranslmed.abb5883
Shi, R., Shan, C., Duan, X., Chen, Z., Liu, P., Song, J., et al. (2020). A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124.
Sissoko, D., Laouenan, C., Folkesson, E., M’Lebing, A. B., Beavogui, A. H., Baize, S., et al. (2016). Experimental treatment with favipiravir for Ebola virus disease (the JIKI Trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med. 13:e1001967. doi: 10.1371/journal.pmed.1001967
Stebbing, J., Krishnan, V., de Bono, S., Ottaviani, S., Casalini, G., Richardson, P. J., et al. (2020). Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 12:e12697.
Stone, J. H., Frigault, M. J., Serling-Boyd, N. J., Fernandes, A. D., Harvey, L., Foulkes, A. S., et al. (2020). Efficacy of tocilizumab in patients hospitalized with Covid-19. N. Engl. J. Med. 383, 2333–2344.
Sun, L., Li, P., Ju, X., Rao, J., Huang, W., Ren, L., et al. (2021). In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell 184, 1865–1883.e20.
Tay, M. Y., Fraser, J. E., Chan, W. K., Moreland, N. J., Rathore, A. P., Wang, C., et al. (2013). Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor ivermectin. Antiviral Res. 99, 301–306. doi: 10.1016/j.antiviral.2013.06.002
Tu, Y. F., Chien, C. S., Yarmishyn, A. A., Lin, Y. Y., Luo, Y. H., Lin, Y. T., et al. (2020). A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 21:2657. doi: 10.3390/ijms21072657
Udwadia, Z. F., Singh, P., Barkate, H., Patil, S., Rangwala, S., Pendse, A., et al. (2021). Efficacy and safety of favipiravir, an oral RNA-dependent RNA polymerase inhibitor, in mild-to-moderate COVID-19: a randomized, comparative, open-label, multicenter, phase 3 clinical trial. Int. J. Infect. Dis. 103, 62–71. doi: 10.1016/j.ijid.2020.11.142
Vankadari, N. (2020). Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents 56:105998. doi: 10.1016/j.ijantimicag.2020.105998
Verdecchia, P., Cavallini, C., Spanevello, A., and Angeli, F. (2020). The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 76, 14–20. doi: 10.1016/j.ejim.2020.04.037
Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., and Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 443, 851–856. doi: 10.1042/bj20120150
Wahl, A., Gralinski, L. E., Johnson, C. E., Yao, W., Kovarova, M., and Dinnon, K. H. III, et al. (2021). SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature 591, 451–457. doi: 10.1038/s41586-021-03312-w
Wang, C., Li, W., Drabek, D., Okba, N. M. A., van Haperen, R., Osterhaus, A., et al. (2020a). A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 11:2251.
Wang, K., Chen, W., Zhang, Z., Deng, Y., Lian, J. Q., Du, P., et al. (2020b). CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 5:283.
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020c). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271. doi: 10.1038/s41422-020-0282-0
Wang, M. Y., Zhao, R., Gao, L. J., Gao, X. F., Wang, D. P., and Cao, J. M. (2020d). SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 10:587269. doi: 10.3389/fcimb.2020.587269
Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., et al. (2020e). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e9.
Wang, Y., Zhou, F., Zhang, D., Zhao, J., Du, R., Hu, Y., et al. (2020f). Evaluation of the efficacy and safety of intravenous remdesivir in adult patients with severe COVID-19: study protocol for a phase 3 randomized, double-blind, placebo-controlled, multicentre trial. Trials 21:422.
Wang, Z., Yang, B., Li, Q., Wen, L., and Zhang, R. (2020g). Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 71, 769–777. doi: 10.1093/cid/ciaa272
Warren, T. K., Jordan, R., Lo, M. K., Ray, A. S., Mackman, R. L., Soloveva, V., et al. (2016). Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531, 381–385.
Wei, R., Zheng, N., Jiang, X., Ma, C., Xu, X., Liu, S., et al. (2020). Early antiviral therapy of abidol combined with lopinavir/ritonavir and recombinant interferon α-2b for patients with COVID-19 in Zhejiang: a multicenter prospective study. Chin. J. Clin. Infect. Dis. 13, 9–15.
Weinreich, D. M., Sivapalasingam, S., Norton, T., Ali, S., Gao, H., Bhore, R., et al. (2021). REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med. 384, 238–251. doi: 10.1056/nejmoa2035002
White, K. M., Rosales, R., Yildiz, S., Kehrer, T., Miorin, L., Moreno, E., et al. (2021). Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 371, 926–931. doi: 10.1126/science.abf4058
WHO Solidarity Trial Consortium, Pan, H., Peto, R., Henao-Restrepo, A. M., Preziosi, M. P., Sathiyamoorthy, V., et al. (2021). Repurposed antiviral drugs for Covid-19 – interim WHO solidarity trial results. N. Engl. J. Med. 384, 497–511. doi: 10.1056/nejmoa2023184
Williamson, B. N., Feldmann, F., Schwarz, B., Meade-White, K., Porter, D. P., Schulz, J., et al. (2020). Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 585, 273–276. doi: 10.1038/s41586-020-2423-5
Woo, P. C., Lau, S. K., Huang, Y., and Yuen, K. Y. (2009). Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 234, 1117–1127. doi: 10.3181/0903-mr-94
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263.
Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020a). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 10, 766–788. doi: 10.1016/j.apsb.2020.02.008
Wu, Y., Li, C., Xia, S., Tian, X., Kong, Y., Wang, Z., et al. (2020b). Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe 27, 891–898.e5.
Wu, Y., Wang, F., Shen, C., Peng, W., Li, D., Zhao, C., et al. (2020c). A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368, 1274–1278. doi: 10.1126/science.abc2241
Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., et al. (2020a). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30, 343–355. doi: 10.1038/s41422-020-0305-x
Xia, S., Zhu, Y., Liu, M., Lan, Q., Xu, W., Wu, Y., et al. (2020b). Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 17, 765–767. doi: 10.1038/s41423-020-0374-2
Xia, W., An, C., Zheng, C., Zhang, J., Huang, M., Wang, Y., et al. (2020c). Clinical observation on 34 patients with novel coronavirus Pneumonia (COVID–19) treated with intergrated traditional Chinese and western medicine. J. Tradit. Chin. Med. 61, 375–382.
Yamamoto, M., Matsuyama, S., Li, X., Takeda, M., Kawaguchi, Y., Inoue, J. I., et al. (2016). Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 60, 6532–6539. doi: 10.1128/aac.01043-16
Yan, Y., Zou, Z., Sun, Y., Li, X., Xu, K. F., Wei, Y., et al. (2013). Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 23, 300–302. doi: 10.1038/cr.2012.165
Yang, L., Liu, W., Yu, X., Wu, M., Reichert, J. M., and Ho, M. (2020). COVID-19 antibody therapeutics tracker: a global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib. Ther. 3, 205–212. doi: 10.1093/abt/tbaa020
Yuan, M., Wu, N. C., Zhu, X., Lee, C. D., So, R. T. Y., Lv, H., et al. (2020). A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633. doi: 10.1126/science.abb7269
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D., and Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820. doi: 10.1056/NEJMoa1211721
Zang, R., Gomez Castro, M. F., McCune, B. T., Zeng, Q., Rothlauf, P. W., Sonnek, N. M., et al. (2020). TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5:eabc3582. doi: 10.1126/sciimmunol.abc3582
Zeng, X., Song, X., Ma, T., Pan, X., Zhou, Y., Hou, Y., et al. (2020). Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res. 19, 4624–4636. doi: 10.1021/acs.jproteome.0c00316
Zeng, Y. M., Xu, X. L., He, X. Q., Tang, S. Q., Li, Y., Huang, Y. Q., et al. (2020). Comparative effectiveness and safety of ribavirin plus interferon-alpha, lopinavir/ritonavir plus interferon-alpha, and ribavirin plus lopinavir/ritonavir plus interferon-alpha in patients with mild to moderate novel coronavirus disease 2019: study protocol. Chin. Med. J. 133, 1132–1134. doi: 10.1097/cm9.0000000000000790
Zhang, D. H., Wu, K. L., Zhang, X., Deng, S. Q., and Peng, B. (2020). In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 18, 152–158. doi: 10.1016/j.joim.2020.02.005
Zhou, J., and Huang, J. (2020). Current findings regarding natural components with potential anti-2019-nCoV activity. Front. Cell Dev. Biol. 8:589. doi: 10.3389/fcell.2020.00589
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020a). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-7
Zhou, W., Liu, Y., Tian, D., Wang, C., Wang, S., Cheng, J., et al. (2020b). Potential benefits of precise corticosteroids therapy for severe 2019-nCoV pneumonia. Signal Transduct. Target. Ther. 5:18.
Zhou, Y., Hou, Y., Shen, J., Huang, Y., Martin, W., and Cheng, F. (2020c). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 6:14.
Zhou, Y., Wang, F., Tang, J., Nussinov, R., and Cheng, F. (2020d). Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health 2, e667–e676.
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733.
Zhu, Z., Lu, Z. H., Xu, T. M., Chen, C., Yang, G., Zha, T., et al. (2020). Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 81, E21–E23.
Keywords: COVID-19, SARS-CoV-2, direct-acting antiviral, host-targeting antiviral, high throughput screening, artificial intelligence, antibody, immuno-regulator
Citation: Mei M and Tan X (2021) Current Strategies of Antiviral Drug Discovery for COVID-19. Front. Mol. Biosci. 8:671263. doi: 10.3389/fmolb.2021.671263
Received: 23 February 2021; Accepted: 08 April 2021;
Published: 13 May 2021.
Edited by:
Tengchuan Jin, University of Science and Technology of China, ChinaReviewed by:
Yuedong Yang, Sun Yat-sen University, ChinaEloise Mastrangelo, National Research Council (CNR), Italy
Copyright © 2021 Mei and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Tan, xutan@tsinghua.edu.cn
 Miao Mei
Miao Mei Xu Tan
Xu Tan