Impaired spatial memory and enhanced long-term potentiation in mice with forebrain-specific ablation of the Stim genes
- 1Program in Neuroscience and Behavioral Disorders, Duke-NUS Graduate Medical School, Singapore, Singapore
- 2Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
- 3Division of Molecular Immunology, Medical Institute of Bioregulation, Kyushu University, Higashi-ku, Japan
- 4Behavioral Neuroscience Laboratory, National Neuroscience Institute, Singapore, Singapore
Recent findings point to a central role of the endoplasmic reticulum-resident STIM (Stromal Interaction Molecule) proteins in shaping the structure and function of excitatory synapses in the mammalian brain. The impact of the Stim genes on cognitive functions remains, however, poorly understood. To explore the function of the Stim genes in learning and memory, we generated three mouse strains with conditional deletion (cKO) of Stim1 and/or Stim2 in the forebrain. Stim1, Stim2, and double Stim1/Stim2 cKO mice show no obvious brain structural defects or locomotor impairment. Analysis of spatial reference memory in the Morris water maze revealed a mild learning delay in Stim1 cKO mice, while learning and memory in Stim2 cKO mice was indistinguishable from their control littermates. Deletion of both Stim genes in the forebrain resulted, however, in a pronounced impairment in spatial learning and memory reflecting a synergistic effect of the Stim genes on the underlying neural circuits. Notably, long-term potentiation (LTP) at CA3-CA1 hippocampal synapses was markedly enhanced in Stim1/Stim2 cKO mice and was associated with increased phosphorylation of the AMPA receptor subunit GluA1, the transcriptional regulator CREB and the L-type Voltage-dependent Ca2+ channel Cav1.2 on protein kinase A (PKA) sites. We conclude that STIM1 and STIM2 are key regulators of PKA signaling and synaptic plasticity in neural circuits encoding spatial memory. Our findings also reveal an inverse correlation between LTP and spatial learning/memory and suggest that abnormal enhancement of cAMP/PKA signaling and synaptic efficacy disrupts the formation of new memories.
Introduction
STIM1 and STIM2 are endoplasmic reticulum (ER)-resident Ca2+ sensors that regulate store-operated Ca2+ entry (SOCE), the major Ca2+ influx pathway in non-excitable cells (Liou et al., 2005; Roos et al., 2005). A recent body of evidence indicates that STIMs also function in the brain and shape synaptic transmission and architecture (Gruszczynska-Biegala et al., 2011; Hartmann et al., 2014; Sun et al., 2014; Garcia-Alvarez et al., 2015). Although both STIM isoforms are expressed throughout the nervous system, STIM1 is more abundant in the cerebellum (Skibinska-Kijek et al., 2009; Hartmann et al., 2014), while STIM2 is the predominant isoform in the hippocampus (Berna-Erro et al., 2009; Skibinska-Kijek et al., 2009; Sun et al., 2014; Garcia-Alvarez et al., 2015), an expression pattern which is consistent with the reported functions of these isoforms in the brain. STIM1 controls mGluR1-dependent synaptic transmission in Purkinje neurons and cerebellar motor behavior (Hartmann et al., 2014), whereas STIM2 influences dendritic spine morphogenesis (Sun et al., 2014; Garcia-Alvarez et al., 2015) and regulates AMPA receptor (AMPAR) phosphorylation and trafficking in the hippocampus (Garcia-Alvarez et al., 2015). Notably, STIM2 has also been associated with brain pathologies. Systemic ablation of the Stim2 gene protects against hypoxic neuronal cell death (Berna-Erro et al., 2009) and reduced levels of STIM2 in a presenilin-1 mouse model of Alzheimer's disease accounts for a decrease in the fraction of mature (mushroom) dendritic spines in hippocampal neurons (Sun et al., 2014).
Most studies on STIM function in the brain report an effect of these genes on neuronal Ca2+ homeostasis (Hartmann et al., 2014; Sun et al., 2014), Ca2+ influx (through SOCE) (Berna-Erro et al., 2009; Sun et al., 2014) or Ca2+ mobilization from intracellular stores (Hartmann et al., 2014), suggesting that the STIM proteins regulate synaptic function primarily through their ability to shape synaptic Ca2+ concentration. Results from our laboratory recently highlighted a novel, SOCE-independent function of STIM2 in excitatory neurons in coupling the AMPAR subunit GluA1 to PKA (Garcia-Alvarez et al., 2015), indicating that STIMs may impact synaptic transmission through multiple signaling pathways. An interesting feature of STIMs is their redistribution to contact sites between the ER and the plasma membrane (PM) in response to lowering of ER Ca2+ (Liou et al., 2005; Roos et al., 2005; Wu et al., 2006) or cAMP elevation in the cytoplasm (Tian et al., 2012; Garcia-Alvarez et al., 2015). Dynamic localization to ER-PM contact sites allow STIMs to interact with and modify the properties of PM channels and receptors both in excitable and non-excitable cells (Park et al., 2009, 2010; Wang et al., 2010; Garcia-Alvarez et al., 2015). Together, these findings highlight an unprecedented level of crosstalk between the ER and the synapse and suggest that ER-to-synapse signaling sculpts functional neural circuits in the brain.
The impact of the Stim genes on behavior remains, however, largely unexplored. Systemic deletion of Stim1 or Stim2 is lethal within days or weeks after birth (Oh-Hora et al., 2008; Berna-Erro et al., 2009), precluding behavioral phenotyping of adult mice. Here, we report the behavioral analysis of Stim1, Stim2, and double Stim1/Stim2 cKO mice, in which the Stim genes are selectively excised from the forebrain after birth. While single gene deletion has no or little effect on cognitive functions, ablation of both Stim genes leads to severe deficits in spatial learning and memory. Surprisingly, double Stim1/Stim2 cKO mice exhibit enhanced LTP and elevated PKA signaling in the hippocampus, suggesting that uncontrolled PKA activity and synaptic strength deleteriously affects the acquisition of spatial memories.
Materials and Methods
Mouse Lines
Stim1fl/fl, Stim2fl/fl, and double Stim1fl/fl/Stim2fl/fl mice, first described in Oh-Hora et al. (2008), were crossed to B6.Cg-Tg(CaMKIIα-Cre)T29-1Stl/J (Tsien et al., 1996a) provided by the Jackson Laboratory (JAX, stock # 005359). The tdTomato Cre reporter line Ai9 (Madisen et al., 2010) was obtained from JAX (stock # 007909). The Stimfl/fl, CaMKIIα-Cre and Ai9 lines are all derived from the C57BL/6J background. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Singapore.
Immunohistochemistry and Confocal Microscopy
Four-month-old mice were anesthetized with pentobarbital and transcardially perfused with phosphate-buffered saline (PBS), followed by 4% paraformaldehyde (PFA) in PBS, pH 7.4. Brains were dissected out, post-fixed in 4% PFA at 4°C overnight and cryoprotected for the next 2 days in 15 and 30% sucrose at 4°C. 40 μm coronal sections were cut on a freezing microtome and stored in cryoprotectant (30% ethylene glycol, 30% glycerol in PBS) at −20°C. For immunostaining, free floating sections were permeabilized with 0.25% Triton X-100 in PBS for 30 min and blocked with 2% BSA, 10% horse serum and 0.25% Triton X-100 in PBS for 1 h at room temperature. Sections were incubated with chicken anti-MAP2 antibody (1:1500, AbCam ab5392) at 4°C overnight, followed by incubation with goat anti-chicken 488 secondary antibody (Alexa Fluor, Life Technologies) for 1 h at room temperature and 5 μM Hoechst 33342 for 15 min. Sections were then mounted on glass slides with 0.2% gelatine in 50 mM Tris-HCl (pH 7.5), and covered with FluorSave (CalBiochem) using coverslips. Slides were imaged on a Zeiss LSM 710 inverted laser scanning confocal microscope using a 10X air objective. The Hoechst and MAP2 channels were excited with 405 and 488 nm solid-state lasers respectively. The full view of coronal sections was acquired using the Tile Scan function of the Zen 2010 B software.
Behavioral Analysis
All behavioral tests were conducted on 3–3.5 month old males. Experiments were performed blind to genotype.
Open Field Test (OFT)
Mice were placed in the middle of a closed arena (45 × 45 cm) and their activity was monitored for 15 min using the ANY-maze™ videotracking system (Stoelting, USA). The following parameters were scored: distance traveled, time in the center, rearing, grooming, and immobility, as previously described in Branchi et al. (2004).
Elevated Plus Maze (EPM)
The gray colored metal alloy maze consists of a 60 cm elevated plus-shaped apparatus with four arms (35 cm length × 5 cm width), two of them surrounded with walls of 15 cm height (Ugo Basile, Italy). Mice were placed on the central platform facing a closed arm and were allowed to freely explore the maze for 5 min. Behavior was automatically analyzed with the ANY-maze™ videotracking system, as described in Bichler et al. (2013).
Morris Water Maze (MWM)
The MWM paradigm was adapted from Manns et al. (2010). Each mouse was placed in a 120 cm diameter gray circular pool filled with 40 cm deep opaque water and containing a fixed hidden (submerged) platform. The animals were tested four times a day for five consecutive days, with an inter-trial of about 20 min. For each trial, mice were released facing the tank wall from one randomly selected starting point (North, South, East, or West), and were allowed to swim until they reach the platform (latency) or for a maximum of 60 s. On the 6th day, a 60 s probe trial was conducted without platform and the time spent in each quadrant was analyzed. Reversal learning was initiated 1 h after the probe trial, and mice were tested 4 times per day for 3 days for their ability to locate the platform in the opposite quadrant. Trials were recorded with a video camera placed above the center of the pool and the analysis was automated through the ANY-maze™ video tracking system.
Radial Arm Maze (RAM)
The RAM task was carried out as previously described (Manns et al., 2010). Mice were maintained at 85% of their free feeding weights. Each of the 8 radial arms (7 × 38 cm) was baited at its extremity with a food pellet. After placing the mouse in the central area (18.5 cm diameter), the trap doors surrounding the center were opened and the animal was allowed to enter one arm, after which all other doors were closed. When the mouse returned to the center, all doors stayed closed during an inter-trial period of 5 s. A trial is completed once the mouse has collected all food pellets. Re-entries into already visited arms are considered working memory errors. Mice were tested daily until they reach 80% correct choices during three consecutive days.
Visual Cliff
The test consists of a transparent plastic box placed in a table with the four edges emerging about 20 cm above the top of the table and a rod running along the middle, separating the box into two sections: one has a checker paper placed on the top surface of the box, whereas the other has the same checker paper placed on the bottom surface of the box. The mouse was placed on the rod and should choose within 5 min to walk on the safe side, where the checker paper is on the top surface of the box or the cliff zone (“fake cliff”) where the checker paper is on the bottom surface of the box. Each mouse is tested for 10 trials with the apparatus turned 180° after five trials. Results are expressed in percentages of chosen zones for each mouse.
Statistics were done using SPSS Software, v18.0. One or Two-Way analysis of variance (ANOVA) followed by Bonferroni post-hoc comparisons was used to compare the mean differences with multiple factors, i.e., between groups (genotypes), and/or intervals (days), as appropriate. Unpaired Student t-test was used when only two groups were compared, unless specified otherwise. Equality of variances was assessed by Levene's Test. Statistical significance was considered only when p < 0.05.
Electrophysiology
After anaesthetization with CO2, male mice (3–4 months old) were decapitated and the brains were quickly removed into artificial cerebrospinal fluid (aCSF) containing (in mM): 124 NaCl, 3.7 KCl, 1.2 KH2PO4, 1 MgSO4·7H2O, 2.5 CaCl2·2H2O, 24.6 NaHCO3, and 10 D-glucose, maintained at 2–4°C. The pH of solution was between 7.3 and 7.4 when bubbled with 95% O2 and 5% CO2. 400 μm-thick transverse hippocampal slices were quickly prepared from the right hippocampus using a manual tissue chopper, transferred onto nylon net in an interface chamber (Scientific Systems Design, Canada) and incubated at 32°C at an aCSF flow rate of 1 ml/min and carbogen consumption of 16 l/h. Slices were incubated for at least 3 h in the chamber before starting the experiments. In all experiments, 2 monopolar, lacquer-coated, stainless-steel electrodes (5 MΩ; AM Systems, Carlsborg) were positioned within the stratum radiatum of the CA1 region; one to stimulate the Schaffer collaterals and another to record the fEPSP responses from the apical dendrites. The signals were amplified by a differential amplifier (Model 1700, AM Systems), digitized and monitored online with a custom software. After the incubation period, an input–output curve (afferent stimulation vs. fEPSP slope) was plotted and the test stimulus intensity for each slice was set to elicit a fEPSP slope 40–50% of its maximal response. Test stimulation consisted of four 0.2 Hz biphasic constant-current pulses (0.1 ms/polarity) delivered every 5 min. In all experiments, at least 30 min of stable baseline was recorded before LTP induction. The strong tetanization protocol (STET) consisted of three trains of high frequency stimulation (each train at 100 Hz, 100 biphasic constant-current pulses, single burst, 0.2 ms pulse duration) delivered with an inter-train interval of 10 min (Sajikumar et al., 2005). Theta-burst stimulation (TBS) consisted of 50 bursts (4 stimuli/burst) at an inter-stimulus interval of 10 ms. The 50 bursts were applied over a period of 30 s at 5 Hz (or at an inter-burst interval of 200 ms) (Huang and Kandel, 2005). The initial slopes of fEPSPs were expressed as percentages of baseline averages. The time-matched, normalized data were averaged across replicate experiments and expressed as mean ± SEM. Statistical analyses were performed with GraphPad Prism 6.0.
Western Blotting
Three month old mouse brains were dissected and homogenized using a Glass/Glass dounce homogenizer. The homogenate was spun at 1000 g to remove cell debris and lysed with RIPA buffer [50 mM Tris-HCl pH = 8, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, complete protease inhibitors, and phosphatases inhibitors (Roche)]. Lysates were then cleared by centrifugation. For immunoblotting 20–40 μg of total protein was loaded. Fractions were then analyzed by SDS-PAGE and immunoblotted using Abs against STIM1 (ProSci, 4119), STIM2 (Cell Signaling, 4917S), GluA1 (Millipore, MAB2263), pSer845-GluA1 (Millipore, 04-1073), CREB (Cell Signaling, 9197), pSer133-CREB (Cell Signaling, 9191), Cav1.2 (Alomone Labs, ACC-003), pSer1928-Cav1.2 (LifeSpan Bioscience, LS-C145147) and GAPDH (Millipore, MAB374). Immunoblots were developed using horseradish peroxidase (HRP)-conjugated secondary Abs (Jackson Laboratories), followed by detection with enhanced chemiluminescence (ECL, Pierce). Exposure was adjusted to prevent saturation of the ECL signal.
Results
Generation of Brain-specific Stim1, Stim2, and Double Stim1/Stim2 cKO Mice
To conditionally inactivate the Stim genes in the forebrain, we crossed mice homozygous for loxP-flanked (floxed, fl) Stim1−, Stim2− or Stim1/Stim2− alleles (known as Stim1fl/fl, Stim2fl/fl, or Stim1fl/fl/Stim2fl/fl) (Oh-Hora et al., 2008) with CaMKIIα-Cre transgenic mice (Tsien et al., 1996a). Floxed Stim males heterozygous for Cre (Stimfl/fl;CaMKIIα-Cre+/−) were then mated with Stimfl/fl;CaMKIIα-Cre−/− females to produce litters consisting of Stimfl/fl;Cre+/− (Stim cKO) and Stimfl/fl;Cre−/− (control littermates). Crossing CaMKIIα-Cre mice to a conditional tdTomato Cre reporter line Ai9 (Madisen et al., 2010) showed that Cre recombinase is turned on around 1 month after birth and is expressed in virtually all CA1 hippocampal neurons after 90 days (Figure S1A in Supplementary Material).
To confirm Cre-mediated excision of the Stim genes in these three cKO strains, we analyzed expression of the STIM proteins in different brain regions harvested from adult animals using isoform-specific Abs against STIM1 and STIM2. STIM1 and STIM2 levels were markedly reduced in Stim1 and Stim2 cKO mice respectively, in both the hippocampus and cortex, when compared to their control littermates (Figure 1A). As expected, no Cre-mediated excision was observed in the cerebellum. Importantly, ablation of a single Stim isoform led to no compensatory increase in the expression of its paralogue. Finally both Stim genes were efficiently knocked out in the double Stim1/Stim2 cKO mouse. Residual expression of the STIM proteins in these cKO lines likely reflects the presence of inhibitory neurons and/or glial cells in the dissected tissues, in which CaMKIIα-Cre is not expressed (Sik et al., 1998). Immunocytochemistry of brain slices revealed no obvious anatomical defects or decrease in neuronal density in Stim1, Stim2 or Stim1/Stim2 cKO mice up to 4 months of age (Figure 1B, we have not analyzed older animals).
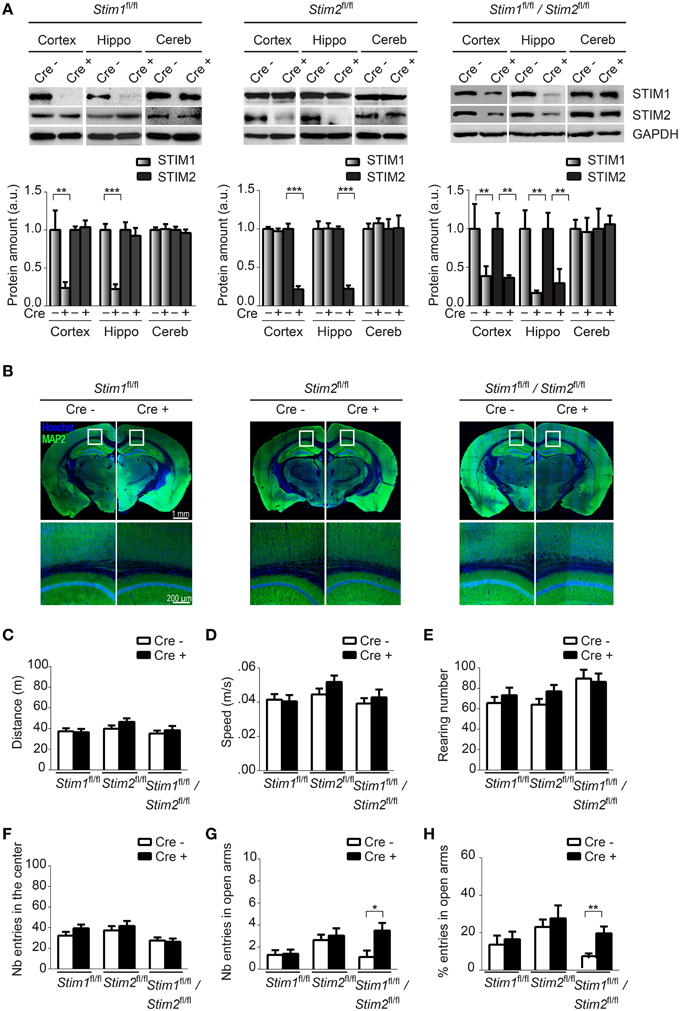
Figure 1. Characterization of forebrain-specific Stim cKO mouse strains. (A) Upper panels. Immunoblots of STIM1 and STIM2 from cortex, hippocampus (hippo), and cerebellum (cereb) harvested from adult Stim1fl/fl;Cre+ (cKO) and Cre− (control), Stim2fl/fl;Cre+ and Cre−, and double Stim1fl/fl/Stim2fl/fl;Cre+ and Cre− mice. Lower panels. Quantification of STIM1 and STIM2 abundance in the Stim lines indicated on top. 6 brains were analyzed for each genotype. Data are normalized to Cre− controls. Blot exposures were adjusted for optimal comparison of Cre− and Cre+ groups and thus do not accurately reflect relative abundance of the STIM isoforms in different brain regions. Means ± SD are shown. **p < 0.01 and ***p < 0.001, t-test. (B) Coronal sections of adult mice (4 months old) immunostained for MAP2 (a neuronal marker) and nuclei (Hoechst). Lower panels are a magnification of the area indicated in the upper panels and show the neuropil in part of the cortex and CA1 region of the hippocampus. (C–F) Locomotor activity probed by the open field test. (C) distance traveled, (D) average speed, (E) rearing, and (F) propensity to visit the center of the field for Stim1fl/fl;Cre+ and Cre−, Stim2fl/fl;Cre+ and Cre− and Stim1fl/fl/Stim2fl/fl;Cre+ and Cre− mice. n = 10 for each group. Means ± SEM are displayed. There is no significant difference across all genotypes. Number (G) and percentage (H) of entries in open arm of the elevated plus maze for Stim1fl/fl;Cre+ (n = 10) and Cre− (n = 10), Stim2fl/fl;Cre+ (n = 22) and Cre− (n = 22) and Stim1fl/fl/Stim2fl/fl;Cre+ (n = 10) and Cre− (n = 10) mice. Means ± SEM are shown. *p < 0.05, **p < 0.01.
Ablation of the Stim Genes does not Affect Locomotor Behavior
To begin to examine the behavior of these Stim cKO mice, we subjected them to the open field test (OFT) which assesses general locomotor activity (Stanford, 2007). We observed no significant difference in distance traveled (Figure 1C), average speed (Figure 1D), rearing (vertical activity, Figure 1E), immobility (Figure S1B) and grooming (Figure S1C) between the three Stim cKO lines and their control littermates or across genotypes. Neither did we detect any changes in time spent in the center (and periphery, data not shown) of the field (Figure 1F), which has been proposed to reflect exploratory and anxiety-related behaviors (Prut and Belzung, 2003). As another popular model for anxiety-like behavior, we used the elevated plus maze (EPM) which is based on rodents' aversion for open spaces (Carobrez and Bertoglio, 2005). The percentage of entries in the closed and open arms was not significantly different in Stim1 and Stim2 cKO mice, relative to their control littermates. The double Stim1/Stim2 cKO mice, however, spent more time in the open arms than their control littermates, suggesting an increased willingness for exploration and reduced anxiety-like behavior (Figures 1G,H). Together, these results reveal no locomotor defects in these three mutant mice and a higher tendency for exploratory behavior (reduced anxiety) in the double Stim1/Stim2 cKO mice apparent in the EPM.
Impaired Spatial Reference Memory in the Double Stim1/Stim2 cKO Mice
We next examined spatial learning and memory in these mutant mice in the Morris water maze (MWM). In this spatial reference memory task, the animal is trained to remember the position of a fixed (hidden) escape platform using allocentric spatial cues. Mutant mice and their control littermates received 4 trials a day for 5 consecutive days and performance was assessed by measuring the time (latency) to find the platform. Control mice (across genotypes) performed equally well in this task and reached the platform within 10 sec by the fifth training day (Figures 2A,D,G,J). Stim1 cKO showed a mild learning defect early on, but performed as well as their control littermates by day 4 (Figures 2A–C). Spatial memory was then analyzed by the probe trial on the 6th day, during which the platform is removed from the pool, and the animal is allowed to swim freely for 60 s. Memory is then inferred based on the time spent in the target quadrant searching for the platform. Both Stim1 cKO mice and control littermates displayed an equally strong bias toward the target quadrant (Figure 2K), indicating that inactivation of the Stim1 gene has no effect on the acquisition of spatial memory. Stim1 cKO mice were also tested for reversal learning, a form of spatial learning which involves extinction of the old platform location and re-learning of its new location. Of interest, Stim1 cKO mice showed delayed reversal learning compared to their control littermates (Figures 2A,B).
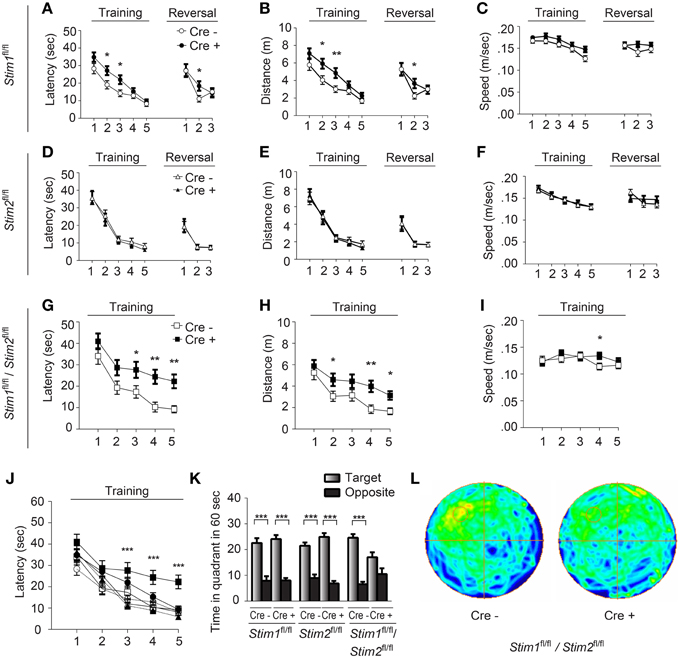
Figure 2. Stim1/Stim2 cKO mice show severe spatial reference memory deficit in the Morris water maze. Latency to find the escape platform (A,D,G,J), distance (B,E,H), and speed (C,F,I) plotted for each training day, or during spatial reversal, for (A–C) Stim1fl/fl;Cre+(n = 18) and Cre− (n = 14), (D–F) Stim2fl/fl;Cre+(n = 10) and Cre− (n = 10), and (G–I) Stim1fl/fl/Stim2fl/fl;Cre+ (n = 10) and Cre− (n = 10) mice. Means ± SEM are shown. *p < 0.05, **p < 0.01, ANOVA comparison for each day. (J) Latency to find platform for each group. Symbols as in (A,D,G). Mean ± SEM are shown. ***p < 0.001, ANOVA. (K,L) Probe trial. (K) Time spent in the target and opposite quadrants. Means ± SEM are shown. ***p < 0.001, Two-Way ANOVA. Only target and opposite quadrants are shown. (L) Group occupancy plot for Stim1fl/fl/Stim2fl/fl;Cre+ and Cre− mice. The platform was located in the upper left quadrant prior to the probe trial.
Analysis of the Stim2 cKO mice in the MWM revealed unaltered performance in spatial learning (Figures 2D–F), probe trials (Figure 2K) and reversal learning (Figures 2D–F). This came as a surprise given the preferential expression of the Stim2 gene in the hippocampus (Berna-Erro et al., 2009; Skibinska-Kijek et al., 2009; Sun et al., 2014; Garcia-Alvarez et al., 2015) and its recent involvement in excitatory synapse structure and function (Sun et al., 2014; Garcia-Alvarez et al., 2015). In marked contrast, double Stim1/Stim2 cKO mice exhibited strong deficits in spatial learning and memory. Although learning was not completely abolished in these animals, it was strongly suppressed compared to their control littermates (Figures 2G–I) or all other genotypes tested (Figure 2J). This severe learning impairment persists until the fifth training day and is associated with poor performance on the probe trial (Figures 2K,L). The time the Stim1/Stim2 cKO mice spent immobile and their average swimming speed during trials were comparable to those of their control littermates (Figure 2I) indicating that this cognitive phenotype is not due to reduced locomotor activity. Likewise, performance in the visual cliff (Figure S2) and behavioral despair tests (i.e., the tail suspension and the forced swim tests, data not shown) argue against a contribution of poor eyesight or lack of motivation to the MWM phenotype. Together these findings show that ablation of both Stim genes in the forebrain results in severe deficits in spatial reference memory. The lack of a clear memory phenotype in single Stim1 or Stim2 cKO mice suggests redundant or synergistic function of the Stim genes in the underlying neural circuits.
Spatial Working Memory is Not Affected in Stim cKO Mutant Mice
We next evaluated the performance of Stim cKO mutant mice in the radial arm maze (RAM), which assesses a form of short-term, hippocampus-dependent, spatial working memory (Olton, 1979). In contrast to the MWM where the target (hidden platform) is fixed throughout the trials, the correct spatial response in the RAM varies within trials and the animal must use newly-acquired spatial information for an optimal reward strategy. Our RAM set-up consists of eight arms radiating from a central platform. Each arm is baited with a food pellet and food rewards are not replaced within a trial. The animals must therefore keep track of each arm they previously visited for efficient performance. Re-entry into a previously baited arm is defined as a working memory error. Performance of Stim1, Stim2, Stim1/Stim2 cKO mice and their control littermates steadily increases to reach a plateau around the 10th trial, at approximately 80% correct choices (about 1.1 error per trial on average). There was no significant difference between Stim cKO mice and their control littermates or across genotypes (Figures 3A–C). Collectively, these findings support the existence of distinct forms of spatial memory (e.g., short-term versus long-term) governed by different neural algorithms (Reisel et al., 2002; Schmitt et al., 2003; Bannerman et al., 2012) and indicate that the Stim genes specifically regulate spatial reference memory.
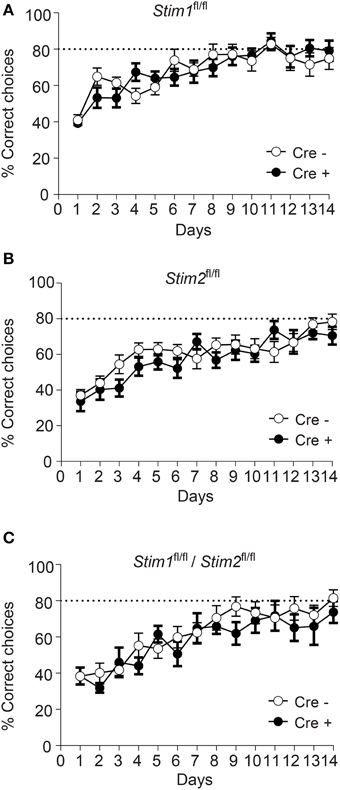
Figure 3. Unaltered spatial working memory in Stim cKO mutant mice. (A–C) Percentage of correct choices in the radial arm maze plotted for each training day as the mean ± SEM. Performance reaches a plateau at 80% correct choices (dashed line) around the 10th day of training. (A) Stim1fl/fl;Cre+ (n = 12) and Cre− (n = 12) mice. (B) Stim2fl/fl;Cre+(n = 12) and Cre− (n = 12) mice. (C) Stim1fl/fl/Stim2fl/fl;Cre+ (n = 7) and Cre− (n = 10) mice. Two Way ANOVA showed a learning effect (p < 0.001 with “days” as within factor) but no genotype effect (p > 0.05, “genotype” as in between factor).
Inactivation of Both Stim Genes Results in Enhanced L-LTP and PKA Signaling
To determine whether the Stim genes regulate synaptic plasticity in hippocampal circuits, we measured long-term potentiation (LTP) at CA3-CA1 hippocampal synapses. LTP was elicited by high-frequency stimulation of the Schaffer collaterals and recorded by measuring field EPSPs (excitatory postsynaptic potentials) in the CA1 region. We chose to elicit LTP using two stimulation paradigms, TBS (theta burst stimulation) and STET (strong tetanus), both of which induce a long-lasting, protein synthesis-dependent form of LTP (late-phase LTP or L-LTP) (Huang and Kandel, 2005; Sajikumar et al., 2005), and we restricted our analysis to the double Stim1/Stim2 cKO mice, since they display spatial memory deficits. To our surprise, both TBS- and STET-induced L-LTP were markedly enhanced in the double Stim1/Stim2 cKO mice compared with their control littermates (Figures 4A,B). Thus, L-LTP and spatial reference memory are inversely correlated in Stim1/Stim2 cKO mice.
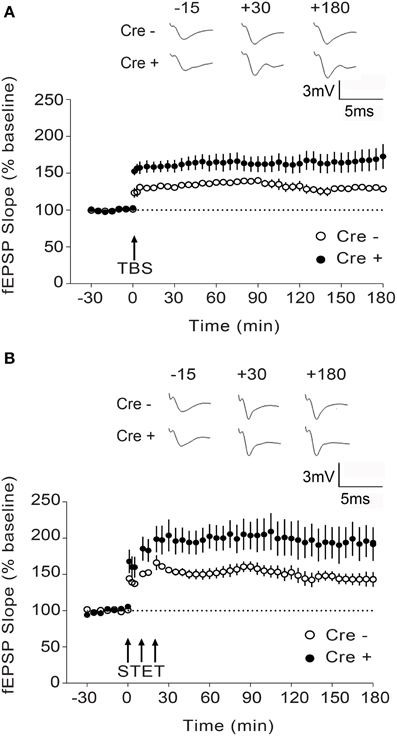
Figure 4. Enhanced L-LTP in Stim1/Stim2 cKO mice. (A) Theta burst (TBS)-induced LTP recorded in CA1 from Stim1/Stim2; Cre+ (n = 8 slices from 5 mice) mice and their control (Cre−) littermates (n = 6 slices from 4 mice). Potentiation in both cases is significantly different from the pre-induction values (+180 min, p < 0.01; paired t-test). By the end of 3 h, mean potentiation of Cre+ group (172.71 ± 16.85%) is significantly different from Cre− group (128.72 ± 3.65%); Two-sample t-test, p < 0.05. (B) Strong tetanization (STET)-induced LTP recorded in CA1 from Stim1/Stim2;Cre+ (n = 6 slices from 4 mice) mice and their control (Cre−) littermates (n = 7 slices from 3 mice). Potentiation in both the groups is significantly different from the pre-induction values (+180 min, p < 0.01; paired t-test). By the end of 3 h, mean potentiation of Cre+ group (194.75 ± 21.13%) is significantly different from Cre− group (143.12 ± 9.16%); Two-sample t-test, p < 0.05. The insets show representative fEPSP traces at −15, +30, and +180 min before and after stimulation. Data are plotted as mean ± SEM.
While most studies report a positive correlation between LTP and spatial memory (reviewed in Silva, 2003), dissociation between LTP and distinct forms of spatial memory has been observed in several different mouse strains (Zamanillo et al., 1999; Kaksonen et al., 2002; Pineda et al., 2004; Rutten et al., 2008). Notably, enhanced LTP and impaired spatial memory has been reported in mice with targeted deletion of genes downregulating adenylate cyclase activity or cAMP levels, resulting in an overall increase in cAMP/PKA signaling (Pineda et al., 2004; Rutten et al., 2008). This, together with data from our own laboratory identifying a central role of STIM2 in cAMP/PKA-dependent phosphorylation of AMPA receptors (Garcia-Alvarez et al., 2015), prompted us to examine PKA signaling in Stim cKO mice. We quantified phosphorylation of three central regulators of synaptic plasticity and memory, namely the AMPAR subunit GluA1, CREB (cAMP response element-binding protein) and the L-type voltage-dependent Ca2+ channel Cav1.2, on serine residues that are known PKA targets (De Jongh et al., 1996; Silva et al., 1998; Esteban et al., 2003).
Phosphorylation of GluA1 (pSer-845), CREB (pSer-133) and Cav1.2 (pSer1928) in the hippocampus was unchanged in Stim1 cKO mice, but was markedly reduced in Stim2 cKO mice (Figures 5A,B), consistent with our previous findings in primary neuron cultures (Garcia-Alvarez et al., 2015). By contrast, phosphorylation of these three proteins was strongly up-regulated in the hippocampus of double Stim1/Stim2 cKO mice. Although, we cannot exclude the possible involvement of other kinases in STIM-dependent regulation of CREB and Cav1.2 phosphorylation (Silva et al., 1998; Yang et al., 2005), reciprocal regulation of GluA1 Ser-845 phosphorylation in Stim2 and Stim1/Stim2 cKO mice is indicative of a complex, synergistic interaction of the STIM isoforms in regulating PKA signaling (see Section Discussion). Increased phosphorylation of GluA1, CREB, and Cav1.2 is associated with elevated activity of these proteins (De Jongh et al., 1996; Silva et al., 1998; Esteban et al., 2003) and is likely to contribute to the enhanced LTP observed in the double Stim1/Stim2 cKO mice. Together, these findings show that the Stim genes are key regulators of phosphorylation and synaptic plasticity in neural circuits underlying spatial memory.
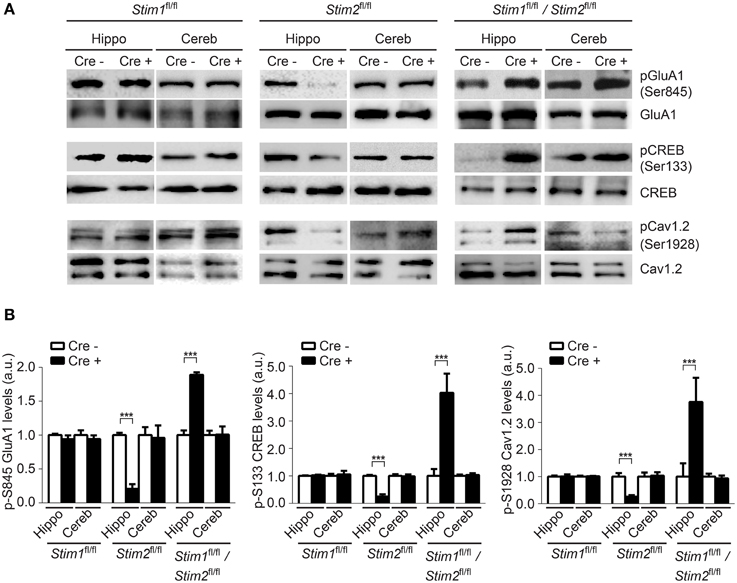
Figure 5. Effect of the Stim genes on PKA signaling. (A) Phospho-immunoblots of pSer845-GluA1, pSer133-CREB, and pSer1928-Cav1.2 in hippocampus (hippo) and cerebellum (cereb) harvested from the three Stim cKO (Cre+) lines and their control (Cre−) littermates. (B) Quantification of pSer845-GluA1, pSer133-CREB, and pSer1928-Cav1.2 levels (n = 3 for each group). Phospho-protein signals are normalized within each Stim genotype to the Cre− group. Blot exposures were adjusted for optimal comparison of phospho-signals in Cre− and Cre+ groups and thus do not accurately reflect relative levels of phosphoproteins in the different Stim lines. Means ± SD are shown. ***p < 0.001, t-test.
Discussion
The Stim Genes are Required for a Specific form of Long-term Spatial Memory
We report here the first behavioral analysis of mutant mice with conditional deletion of the Stim genes in the forebrain. Stim2 cKO mice show no apparent defect in spatial reference memory (MWM), spatial working memory (RAM) or in any of the other behavioral tests we conducted. These results are in apparent contradiction with an earlier study reporting impaired performance of Stim2−/− null mice in the MWM (Berna-Erro et al., 2009). Stim2−/− null mice, however, die after 8 weeks (Berna-Erro et al., 2009) or even earlier (4–5 weeks, Oh-Hora et al., 2008), which complicates the interpretation of learning/memory tasks, as poor performance in these tasks could result from pleiotropic effects on development/health, rather than a specific function of the Stim2 gene in circuits encoding memory.
Forebrain-specific inactivation of Stim1 has a subtle impact on spatial learning and memory. Spatial learning is delayed in the Stim1 cKO mice in early trials of the MWM, although by the fifth trial, performance is similar to their control littermates. In addition, spatial reversal in the MWM is impaired in these mutant mice, a phenotype recently described in a mouse strain with conditional ablation of the NMDA receptor subunit Grin1 (GluN1) in the dentate gyrus and CA1 regions of the hippocampus (Bannerman et al., 2012). Decreased performance in spatial reversal reflects an inability to learn a new location in a familiar environment and distinguish between two competing or overlapping memories, a phenomenon akin to pattern separation (Bannerman et al., 2014).
Inactivation of both Stim genes in the forebrain resulted in a severe deficit in spatial reference memory in the MWM. Memory impairment in the double Stim1/Stim2 cKO mice is comparable in extent to that seen in floxed GluN1 mice crossed to the same Cre driver line (Tsien et al., 1996b). This, together with the absence of any detectable anatomical defects in Stim1/Stim2 cKO mice suggests that the STIM proteins regulate key aspects of synaptic function/plasticity in neural circuits encoding spatial memory.
Of interest, ablation of both Stim genes has no effect on spatial working memory (RAM) suggesting a specific impact of the Stim genes on neural circuits encoding spatial reference memory. Dissociation between spatial reference memory and spatial working memory has been observed before, in mouse strains with genetic ablation of glutamate receptors. GluA1 null mice show impaired spatial working memory in the RAM (Reisel et al., 2002) but exhibit intact spatial reference memory in the MWM. Similarly, mice with targeted deletion of GluN1 in dentate gyrus/CA1 show selective impairment of spatial working memory (Bannerman et al., 2012). Our findings therefore support the idea that spatial memory is not a single process, but instead, takes on distinct forms that can be genetically disentangled.
Dual Role of the Stim Genes in Cognition and Emotion
Impaired performance of Stim1/Stim2 cKO mice in the MWM and their reduced anxiety-like behavior in the EPM suggest that the Stim genes impact circuits regulating both cognition and emotion. Lesion studies, gene expression analyses and anatomical connectivity of the hippocampus have led to a model describing its functional parcellation into at least two sub-regions (Fanselow and Dong, 2010). The dorsal hippocampus (DH) appears to be specifically involved in spatial and declarative memories (Moser et al., 1995), while the ventral hippocampus (VH) seems to preferentially control emotional behaviors, in particular those associated with negative affect, like anxiety or fear (Kjelstrup et al., 2002; Maren and Holt, 2004). Of interest, lesion of the VH (Mchugh et al., 2004), or inactivation of the NMDA receptor subunit GluN1 in the dentate gyrus (Niewoehner et al., 2007) both result in increased time spent in the open (anxiogenic) arms of the EPM, a phenotype that we also observed in the Stim1/Stim2 cKO mice. Based on these findings and the expression of Stim1 and Stim2 throughout the dorso-lateral axis of the hippocampus (The Allen mouse brain Atlas, Lein et al., 2007), we speculate that the Stim genes regulate both dorsal and ventral functions of the hippocampus.
LTP and Spatial Memory
LTP (or an LTP-like process) is widely considered as the dominant cellular mechanism underlying experience-dependent changes in synaptic connections and information storage. Many of its characteristics (associativity, cooperativity, and temporal resilience) are consistent with a role in memory formation and maintenance. In addition, a massive body of literature (reviewed in Silva, 2003) suggests that LTP and hippocampus-dependent forms of memory rely on the same cellular pathways and correlate with one another. For example, ablation of the GluN1 subunit of the NMDAR in the forebrain obliterates both LTP and spatial memory (Tsien et al., 1996b), while overexpression of GluN2B has the opposite effect (Tang et al., 1999). This has led to the idea that manipulations boosting LTP may have therapeutic potential for cognitive enhancement. However, an increasing number of studies, relying on different genetically-modified mouse strains, also report clear dissociation of LTP and memory, thereby challenging the causal relationship between these two processes (Meiri et al., 1998; Migaud et al., 1998; Zamanillo et al., 1999; Pineda et al., 2004; Rutten et al., 2008; Kim et al., 2009; Dilekoz et al., 2015). Relevant to our work, genetic manipulations of key plasticity-regulating genes, such as PSD-95 (Migaud et al., 1998), IRSp53 (Kim et al., 2009), PTPδ (Uetani et al., 2000), and components of the cAMP signaling pathway (Pineda et al., 2004; Bourtchouladze et al., 2006; Rutten et al., 2008; Viosca et al., 2009) have been shown to impair spatial memory while, at the same time, enhance LTP. Our findings therefore further support the view that enhancement of LTP in the hippocampus does not necessarily translate into superior learning and memory functions. One possible interpretation of these data is that abnormally high synaptic potentiation alters synaptic plasticity and disrupts input-specific learning rules in the hippocampus (see Pineda et al., 2004). In support of this model, enhanced LTP results in abnormal place cell activity (Taverna et al., 2005) and saturation of LTP impairs spatial learning (Moser et al., 1998). Additionally, mathematical simulations of neural networks indicate that synaptic potentiation must be proportionally constrained by synaptic depression for optimal information storage (Sejnowski, 1977; Dayan and Willshaw, 1991; Migaud et al., 1998), suggesting that uncontrolled LTP may negatively impact network performance.
Synergistic Regulation of PKA Signaling by the Stim Genes
cAMP/PKA signaling regulates multiple aspects of neuronal and synaptic plasticity and is crucial for learning and memory in both invertebrates and vertebrates (reviewed in Silva et al., 1998; Kandel, 2001). Proper dosage of cAMP signaling appears to be central for information storage, as genetic mutations with both negative and positive effects on this signal transduction pathway can be detrimental to memory formation (Livingstone et al., 1984; Skoulakis et al., 1993; Pineda et al., 2004).
We have previously demonstrated an essential role of STIM2 in mediating PKA phosphorylation of GluA1 (Garcia-Alvarez et al., 2015). Results described in the present study confirm involvement of the Stim genes in regulating phosphorylation of PKA targets in vivo, but also reveal a complex interaction of these genes in controlling PKA signaling in the hippocampus. Indeed, PKA phosphorylation of GluA1 on Ser-845 is markedly upregulated in Stim1/Stim2 cKO mice, but is instead repressed or unaffected in Stim2 and Stim1 cKO mice respectively. The same reciprocal pattern is observed for pSer1928-Cav1.2 and pSer133-CREB, although it is possible that STIMs regulate phosphorylation of these sites through kinases other than PKA (Silva et al., 1998; Yang et al., 2005).
The impact of single and double gene ablation on PKA signaling implies the presence of synergy between the Stim genes (i.e., the contribution of both gene deletions to the PKA phenotype far exceeds the expected additive effects of individual gene deletions). A recent survey suggests that synergy often arises from functionally redundant gene pairs, particularly among signaling genes, providing increased adaptability, robustness and evolvability (Kafri et al., 2009). Such a synergistic interaction between the Stim genes could be important for fine-tuning PKA activity and synaptic plasticity.
The effect of the Stim genes on phosphorylation of GluA1, CREB and Cav1.2 is likely to influence synaptic plasticity and behavior in multiple ways. PKA phosphorylation of GluA1 on Ser-845 promotes surface delivery and synaptic recruitment of the AMPAR during LTP (Esteban et al., 2003; Oh et al., 2006). Increased pSer-845 of GluA1 in Stim1/Stim2 cKO mice is thus likely to contribute to the observed enhancement in the induction of LTP. Similarly, elevated levels of pSer1928-Cav1.2 and pSer133-CREB could promote L-LTP in the Stim1/Stim2 cKO mice, as both Cav1.2 and CREB are necessary for protein synthesis-dependent LTP in the hippocampus and long-term spatial memory (Bourtchuladze et al., 1994; Moosmang et al., 2005).
In addition to the proposed role of the STIM proteins in regulating PKA signaling outputs, STIMs are also critical regulators of Ca2+ homeostasis. The aberrant synaptic potentiation and behavior observed in the double Stim1/Stim2 cKO mice could therefore also be linked to altered ER Ca2+ release (Hartmann et al., 2014) or Ca2+ influx (Sun et al., 2014) at the synapse.
In conclusion, we identified novel functions of the Stim genes in shaping excitatory circuits underlying spatial memory in the mouse brain. These findings, together with a handful of recent papers (Hartmann et al., 2014; Sun et al., 2014; Garcia-Alvarez et al., 2015) point to an emerging role of ER-to-synapse signaling in the mammalian nervous system, and are of immediate relevance to synaptic disorders associated with ER dysfunction.
Author Contributions
ZB and GG performed the behavioral analyses. MS performed the electrophysiology experiments. GG, BL, and KY did the biochemistry and immunohistochemistry experiments. MO generated the Stim cKO mice. ZB, SS, and MF designed and supervised the project. MF wrote the manuscript.
Conflict of Interest Statement
This work is partly funded by a joint grant from the Agency of Science Technology and Research (Astar, Singapore) and Janssen Pharmaceuticals, Inc. Janssen Pharmaceuticals had no involvement in any aspect of study design, data collection, analysis and interpretation, or decision to submit this manuscript for publication. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tan Li Ting, Rahim Dini, and Toh Lian Aik for their excellent technical assistance and all members of the MF, ZB, and SS labs for critical inputs. This work was supported by grants to MF from the Singapore Ministry of Education Academic Research Fund (MOE2011-T2-1-107), and the Agency for Science, Technoloy and Research (Astar/JJSI 13/1/24/26/011). SS is supported by the National Medical Research Council, Singapore (NMRC-CBRG-0041/2013). ZB is supported by the Singhealth Foundation (SHF/FG504P/2012) and Singapore Millenium Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fnbeh.2015.00180
Figure S1. (A) tdTomato expression in CA1 pyramidal neurons in the tdTomato Cre reporter line (Ai9) crossed to CaMKIIa-Cre+ mice, atP30 and P90. Nuclei were stained with Hoechst. (B) Immobility and (C) grooming data derived from the open field test for each of the Stim cKO lines and control littermates.
Figure S2. Visual cliff. Percentage of safe actions (correct responses) in the cliff test after 10 trials (mean ± SEM) in Stim1/Stim2;Cre+ (cKO) mice (n = 7) and control (Cre−) littermates (n = 10). There was no significant difference between the two groups.
Abbreviations
STIM, Stromal Interaction Molecule; LTP, long-term potentiation; CREB, cAMP response elementbinding protein; PKA, protein kinase A; AMPA, α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; ER, endoplasmic reticulum; SOCE, store-operated Ca2+ entry; OFT, Open Field Test; EPM, Elevated Plus Maze; MWM, Morris Water Maze; RAM, Radial ArmMaze; GluA1,AMPA-type glutamate receptor subunit 1; GluN1,NMDA-type glutamate receptor subunit 1.
References
Bannerman, D. M., Bus, T., Taylor, A., Sanderson, D. J., Schwarz, I., Jensen, V., et al. (2012). Dissecting spatial knowledge from spatial choice by hippocampal NMDA receptor deletion. Nat. Neurosci. 15, 1153–1159. doi: 10.1038/nn.3166
Bannerman, D. M., Sprengel, R., Sanderson, D. J., Mchugh, S. B., Rawlins, J. N., Monyer, H., et al. (2014). Hippocampal synaptic plasticity, spatial memory and anxiety. Nat. Rev. Neurosci. 15, 181–192. doi: 10.1038/nrn3677
Berna-Erro, A., Braun, A., Kraft, R., Kleinschnitz, C., Schuhmann, M. K., Stegner, D., et al. (2009). STIM2 regulates capacitive Ca2+ entry in neurons and plays a key role in hypoxic neuronal cell death. Sci. Signal. 2, ra67. doi: 10.1126/scisignal.2000522
Bichler, Z., Lim, H. C., Zeng, L., and Tan, E. K. (2013). Non-motor and motor features in LRRK2 transgenic mice. PLoS ONE 8:e70249. doi: 10.1371/journal.pone.0070249
Bourtchouladze, R., Patterson, S. L., Kelly, M. P., Kreibich, A., Kandel, E. R., and Abel, T. (2006). Chronically increased Gsalpha signaling disrupts associative and spatial learning. Learn. Mem. 13, 745–752. doi: 10.1101/lm.354106
Bourtchuladze, R., Frenguelli, B., Blendy, J., Cioffi, D., Schutz, G., and Silva, A. J. (1994). Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68. doi: 10.1016/0092-8674(94)90400-6
Branchi, I., Bichler, Z., Minghetti, L., Delabar, J. M., Malchiodi-Albedi, F., Gonzalez, M. C., et al. (2004). Transgenic mouse in vivo library of human Down syndrome critical region 1: association between DYRK1A overexpression, brain development abnormalities, and cell cycle protein alteration. J. Neuropathol. Exp. Neurol. 63, 429–440.
Carobrez, A. P., and Bertoglio, L. J. (2005). Ethological and temporal analyses of anxiety-like behavior: the elevated plus-maze model 20 years on. Neurosci. Biobehav. Rev. 29, 1193–1205. doi: 10.1016/j.neubiorev.2005.04.017
Dayan, P., and Willshaw, D. J. (1991). Optimising synaptic learning rules in linear associative memories. Biol. Cybern. 65, 253–265. doi: 10.1007/BF00206223
De Jongh, K. S., Murphy, B. J., Colvin, A. A., Hell, J. W., Takahashi, M., and Catterall, W. A. (1996). Specific phosphorylation of a site in the full-length form of the alpha 1 subunit of the cardiac L-type calcium channel by adenosine 3',5'-cyclic monophosphate-dependent protein kinase. Biochemistry 35, 10392–10402. doi: 10.1021/bi953023c
Dilekoz, E., Houben, T., Eikermann-Haerter, K., Balkaya, M., Lenselink, A. M., Whalen, M. J., et al. (2015). Migraine mutations impair hippocampal learning despite enhanced long-term potentiation. J. Neurosci. 35, 3397–3402. doi: 10.1523/JNEUROSCI.2630-14.2015
Esteban, J. A., Shi, S. H., Wilson, C., Nuriya, M., Huganir, R. L., and Malinow, R. (2003). PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat. Neurosci. 6, 136–143. doi: 10.1038/nn997
Fanselow, M. S., and Dong, H. W. (2010). Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 65, 7–19. doi: 10.1016/j.neuron.2009.11.031
Garcia-Alvarez, G., Lu, B., Yap, K. A., Wong, L. C., Thevathasan, J. V., Lim, L., et al. (2015). STIM2 regulates PKA-dependent phosphorylation and trafficking of AMPARs. Mol. Biol. Cell. 26, 1141–1159. doi: 10.1091/mbc.E14-07-1222
Gruszczynska-Biegala, J., Pomorski, P., Wisniewska, M. B., and Kuznicki, J. (2011). Differential roles for STIM1 and STIM2 in store-operated calcium entry in rat neurons. PLoS ONE 6:e19285. doi: 10.1371/journal.pone.0019285
Hartmann, J., Karl, R. M., Alexander, R. P., Adelsberger, H., Brill, M. S., Ruhlmann, C., et al. (2014). STIM1 controls neuronal Ca(2+) signaling, mGluR1-Dependent Synaptic Transmission, and cerebellar motor behavior. Neuron 82, 635–644. doi: 10.1016/j.neuron.2014.03.027
Huang, Y. Y., and Kandel, E. R. (2005). Theta frequency stimulation up-regulates the synaptic strength of the pathway from CA1 to subiculum region of hippocampus. Proc. Natl. Acad. Sci. U.S.A. 102, 232–237. doi: 10.1073/pnas.0408368102
Kafri, R., Springer, M., and Pilpel, Y. (2009). Genetic redundancy: new tricks for old genes. Cell 136, 389–392. doi: 10.1016/j.cell.2009.01.027
Kaksonen, M., Pavlov, I., Voikar, V., Lauri, S. E., Hienola, A., Riekki, R., et al. (2002). Syndecan-3-deficient mice exhibit enhanced LTP and impaired hippocampus-dependent memory. Mol. Cell. Neurosci. 21, 158–172. doi: 10.1006/mcne.2002.1167
Kandel, E. R. (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030–1038. doi: 10.1126/science.1067020
Kim, M. H., Choi, J., Yang, J., Chung, W., Kim, J. H., Paik, S. K., et al. (2009). Enhanced NMDA receptor-mediated synaptic transmission, enhanced long-term potentiation, and impaired learning and memory in mice lacking IRSp53. J. Neurosci. 29, 1586–1595. doi: 10.1523/JNEUROSCI.4306-08.2009
Kjelstrup, K. G., Tuvnes, F. A., Steffenach, H. A., Murison, R., Moser, E. I., and Moser, M. B. (2002). Reduced fear expression after lesions of the ventral hippocampus. Proc. Natl. Acad. Sci. U.S.A. 99, 10825–10830. doi: 10.1073/pnas.152112399
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. doi: 10.1038/nature05453
Liou, J., Kim, M. L., Heo, W. D., Jones, J. T., Myers, J. W., Ferrell, J. E. Jr., et al. (2005). STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235–1241. doi: 10.1016/j.cub.2005.05.055
Livingstone, M. S., Sziber, P. P., and Quinn, W. G. (1984). Loss of calcium/calmodulin responsiveness in adenylate cyclase of rutabaga, a Drosophila learning mutant. Cell 37, 205–215. doi: 10.1016/0092-8674(84)90316-7
Madisen, L., Zwingman, T. A., Sunkin, S. M., Oh, S. W., Zariwala, H. A., Gu, H., et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. doi: 10.1038/nn.2467
Manns, M., Bichler, Z., Leske, O., and Heumann, R. (2010). Neuronal Ras activation inhibits adult hippocampal progenitor cell division and impairs spatial short-term memory. Genes Brain Behav. 9, 525–536. doi: 10.1111/j.1601-183x.2010.00584.x
Maren, S., and Holt, W. G. (2004). Hippocampus and Pavlovian fear conditioning in rats: muscimol infusions into the ventral, but not dorsal, hippocampus impair the acquisition of conditional freezing to an auditory conditional stimulus. Behav. Neurosci. 118, 97–110. doi: 10.1037/0735-7044.118.1.97
Mchugh, S. B., Deacon, R. M., Rawlins, J. N., and Bannerman, D. M. (2004). Amygdala and ventral hippocampus contribute differentially to mechanisms of fear and anxiety. Behav. Neurosci. 118, 63–78. doi: 10.1037/0735-7044.118.1.63
Meiri, N., Sun, M. K., Segal, Z., and Alkon, D. L. (1998). Memory and long-term potentiation (LTP) dissociated: normal spatial memory despite CA1 LTP elimination with Kv1.4 antisense. Proc. Natl. Acad. Sci. U.S.A. 95, 15037–15042. doi: 10.1073/pnas.95.25.15037
Migaud, M., Charlesworth, P., Dempster, M., Webster, L. C., Watabe, A. M., Makhinson, M., et al. (1998). Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396, 433–439. doi: 10.1038/24790
Moosmang, S., Haider, N., Klugbauer, N., Adelsberger, H., Langwieser, N., Muller, J., et al. (2005). Role of hippocampal Cav1.2 Ca2+ channels in NMDA receptor-independent synaptic plasticity and spatial memory. J. Neurosci. 25, 9883–9892. doi: 10.1523/JNEUROSCI.1531-05.2005
Moser, E. I., Krobert, K. A., Moser, M. B., and Morris, R. G. (1998). Impaired spatial learning after saturation of long-term potentiation. Science 281, 2038–2042. doi: 10.1126/science.281.5385.2038
Moser, M. B., Moser, E. I., Forrest, E., Andersen, P., and Morris, R. G. (1995). Spatial learning with a minislab in the dorsal hippocampus. Proc. Natl. Acad. Sci. U.S.A. 92, 9697–9701. doi: 10.1073/pnas.92.21.9697
Niewoehner, B., Single, F. N., Hvalby, O., Jensen, V., Meyer Zum Alten Borgloh, S., Seeburg, P. H., et al. (2007). Impaired spatial working memory but spared spatial reference memory following functional loss of NMDA receptors in the dentate gyrus. Eur. J. Neurosci. 25, 837–846. doi: 10.1111/j.1460-9568.2007.05312.x
Oh, M. C., Derkach, V. A., Guire, E. S., and Soderling, T. R. (2006). Extrasynaptic membrane trafficking regulated by GluR1 serine 845 phosphorylation primes AMPA receptors for long-term potentiation. J. Biol. Chem. 281, 752–758. doi: 10.1074/jbc.M509677200
Oh-Hora, M., Yamashita, M., Hogan, P. G., Sharma, S., Lamperti, E., Chung, W., et al. (2008). Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432–443. doi: 10.1038/ni1574
Olton, D. S. (1979). Mazes, maps, and memory. Am. Psychol. 34, 583–596. doi: 10.1037/0003-066X.34.7.583
Park, C. Y., Hoover, P. J., Mullins, F. M., Bachhawat, P., Covington, E. D., Raunser, S., et al. (2009). STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136, 876–890. doi: 10.1016/j.cell.2009.02.014
Park, C. Y., Shcheglovitov, A., and Dolmetsch, R. (2010). The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science 330, 101–105. doi: 10.1126/science.1191027
Pineda, V. V., Athos, J. I., Wang, H., Celver, J., Ippolito, D., Boulay, G., et al. (2004). Removal of G(ialpha1) constraints on adenylyl cyclase in the hippocampus enhances LTP and impairs memory formation. Neuron 41, 153–163. doi: 10.1016/S0896-6273(03)00813-4
Prut, L., and Belzung, C. (2003). The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. Eur. J. Pharmacol. 463, 3–33. doi: 10.1016/S0014-2999(03)01272-X
Reisel, D., Bannerman, D. M., Schmitt, W. B., Deacon, R. M., Flint, J., Borchardt, T., et al. (2002). Spatial memory dissociations in mice lacking GluR1. Nat. Neurosci. 5, 868–873. doi: 10.1038/nn910
Roos, J., Digregorio, P. J., Yeromin, A. V., Ohlsen, K., Lioudyno, M., Zhang, S., et al. (2005). STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169, 435–445. doi: 10.1083/jcb.200502019
Rutten, K., Misner, D. L., Works, M., Blokland, A., Novak, T. J., Santarelli, L., et al. (2008). Enhanced long-term potentiation and impaired learning in phosphodiesterase 4D-knockout (PDE4D) mice. Eur. J. Neurosci. 28, 625–632. doi: 10.1111/j.1460-9568.2008.06349.x
Sajikumar, S., Navakkode, S., and Frey, J. U. (2005). Protein synthesis-dependent long-term functional plasticity: methods and techniques. Curr. Opin. Neurobiol. 15, 607–613. doi: 10.1016/j.conb.2005.08.009
Schmitt, W. B., Deacon, R. M., Seeburg, P. H., Rawlins, J. N., and Bannerman, D. M. (2003). A within-subjects, within-task demonstration of intact spatial reference memory and impaired spatial working memory in glutamate receptor-A-deficient mice. J. Neurosci. 23, 3953–3959.
Sejnowski, T. J. (1977). Statistical constraints on synaptic plasticity. J. Theor. Biol. 69, 385–389. doi: 10.1016/0022-5193(77)90146-1
Sik, A., Hajos, N., Gulacsi, A., Mody, I., and Freund, T. F. (1998). The absence of a major Ca2+ signaling pathway in GABAergic neurons of the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 95, 3245–3250. doi: 10.1073/pnas.95.6.3245
Silva, A. J. (2003). Molecular and cellular cognitive studies of the role of synaptic plasticity in memory. J. Neurobiol. 54, 224–237. doi: 10.1002/neu.10169
Silva, A. J., Kogan, J. H., Frankland, P. W., and Kida, S. (1998). CREB and memory. Annu. Rev. Neurosci. 21, 127–148. doi: 10.1146/annurev.neuro.21.1.127
Skibinska-Kijek, A., Wisniewska, M. B., Gruszczynska-Biegala, J., Methner, A., and Kuznicki, J. (2009). Immunolocalization of STIM1 in the mouse brain. Acta Neurobiol. Exp. (Wars). 69, 413–428.
Skoulakis, E. M., Kalderon, D., and Davis, R. L. (1993). Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron 11, 197–208. doi: 10.1016/0896-6273(93)90178-T
Stanford, S. C. (2007). The open field test: reinventing the wheel. J. Psychopharmacol. 21, 134–135. doi: 10.1177/0269881107073199
Sun, S., Zhang, H., Liu, J., Popugaeva, E., Xu, N. J., Feske, S., et al. (2014). Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice. Neuron 82, 79–93. doi: 10.1016/j.neuron.2014.02.019
Tang, Y. P., Shimizu, E., Dube, G. R., Rampon, C., Kerchner, G. A., Zhuo, M., et al. (1999). Genetic enhancement of learning and memory in mice. Nature 401, 63–69. doi: 10.1038/43432
Taverna, F. A., Georgiou, J., Mcdonald, R. J., Hong, N. S., Kraev, A., Salter, M. W., et al. (2005). Defective place cell activity in nociceptin receptor knockout mice with elevated NMDA receptor-dependent long-term potentiation. J Physiol. 565, 579–591. doi: 10.1113/jphysiol.2004.081802
Tian, G., Tepikin, A. V., Tengholm, A., and Gylfe, E. (2012). cAMP induces stromal interaction molecule 1 (STIM1) puncta but neither Orai1 protein clustering nor store-operated Ca2+ entry (SOCE) in islet cells. J. Biol. Chem. 287, 9862–9872. doi: 10.1074/jbc.M111.292854
Tsien, J. Z., Chen, D. F., Gerber, D., Tom, C., Mercer, E. H., Anderson, D. J., et al. (1996a). Subregion- and cell type-restricted gene knockout in mouse brain. Cell 87, 1317–1326. doi: 10.1016/S0092-8674(00)81826-7
Tsien, J. Z., Huerta, P. T., and Tonegawa, S. (1996b). The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell 87, 1327–1338. doi: 10.1016/S0092-8674(00)81827-9
Uetani, N., Kato, K., Ogura, H., Mizuno, K., Kawano, K., Mikoshiba, K., et al. (2000). Impaired learning with enhanced hippocampal long-term potentiation in PTPdelta-deficient mice. EMBO J. 19, 2775–2785. doi: 10.1093/emboj/19.12.2775
Viosca, J., Malleret, G., Bourtchouladze, R., Benito, E., Vronskava, S., Kandel, E. R., et al. (2009). Chronic enhancement of CREB activity in the hippocampus interferes with the retrieval of spatial information. Learn. Mem. 16, 198–209. doi: 10.1101/lm.1220309
Wang, Y., Deng, X., Mancarella, S., Hendron, E., Eguchi, S., Soboloff, J., et al. (2010). The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science 330, 105–109. doi: 10.1126/science.1191086
Wu, M. M., Buchanan, J., Luik, R. M., and Lewis, R. S. (2006). Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174, 803–813. doi: 10.1083/jcb.200604014
Yang, L., Liu, G., Zakharov, S. I., Morrow, J. P., Rybin, V. O., Steinberg, S. F., et al. (2005). Ser1928 is a common site for Cav1.2 phosphorylation by protein kinase C isoforms. J. Biol. Chem. 280, 207–214. doi: 10.1074/jbc.M410509200
Keywords: excitatory synapse, AMPA receptor, endoplasmic reticulum, PKA, spatial memory, long-term potentiation, STIM1 and STIM2
Citation: Garcia-Alvarez G, Shetty MS, Lu B, Yap KAF, Oh-Hora M, Sajikumar S, Bichler Z and Fivaz M (2015) Impaired spatial memory and enhanced long-term potentiation in mice with forebrain-specific ablation of the Stim genes. Front. Behav. Neurosci. 9:180. doi: 10.3389/fnbeh.2015.00180
Received: 13 April 2015; Accepted: 29 June 2015;
Published: 14 July 2015.
Edited by:
Gal Richter-Levin, University of Haifa, IsraelReviewed by:
Phillip R. Zoladz, Ohio Northern University, USAMartín Cammarota, Federal University of Rio Grande do Norte, Brazil
Copyright © 2015 Garcia-Alvarez, Shetty, Lu, Yap, Oh-Hora, Sajikumar, Bichler and Fivaz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zoë Bichler, Behavioral Neuroscience Laboratory, National Neuroscience Institute, 11 Jalan Tan Tock Seng, Singapore 308433, Singapore, zoe_bichler@nni.com.sg;
Marc Fivaz, Program in Neuroscience and Behavioral Disorders, Duke-NUS Graduate Medical School, 8 College Road, Singapore 169857, Singapore, marc.fivaz@duke-nus.edu.sg
 Gisela Garcia-Alvarez
Gisela Garcia-Alvarez Mahesh S. Shetty
Mahesh S. Shetty Bo Lu
Bo Lu Kenrick An Fu Yap
Kenrick An Fu Yap Masatsugu Oh-Hora
Masatsugu Oh-Hora Sreedharan Sajikumar
Sreedharan Sajikumar Zoë Bichler
Zoë Bichler Marc Fivaz
Marc Fivaz