- 1Department of Plant Agriculture, University of Guelph, Guelph, ON, Canada
- 2Department of Pharmacognosy, Faculty of Pharmacy, Mansoura University, Mansoura, Egypt
The plant is an attractive versatile home for diverse associated microbes. A subset of these microbes produces a diversity of anti-microbial natural products including polyketides, non-ribosomal peptides, terpenoids, heterocylic nitrogenous compounds, volatile compounds, bacteriocins, and lytic enzymes. In recent years, detailed molecular analysis has led to a better understanding of the underlying genetic mechanisms. New genomic and bioinformatic tools have permitted comparisons of orthologous genes between species, leading to predictions of the associated evolutionary mechanisms responsible for diversification at the genetic and corresponding biochemical levels. The purpose of this review is to describe the biodiversity of biosynthetic genes of plant-associated bacteria and fungi that encode selected examples of antimicrobial natural products. For each compound, the target pathogen and biochemical mode of action are described, in order to draw attention to the complexity of these phenomena. We review recent information of the underlying molecular diversity and draw lessons through comparative genomic analysis of the orthologous coding sequences (CDS). We conclude by discussing emerging themes and gaps, discuss the metabolic pathways in the context of the phylogeny and ecology of their microbial hosts, and discuss potential evolutionary mechanisms that led to the diversification of biosynthetic gene clusters.
Introduction
The plant is an attractive versatile home for diverse microbes that can colonize internal plant tissues (endophytes), live on the surface (epiphytes) or in the soil surrounding the root system (rhizosphere microbiota) (Barea et al., 2005; Johnston-Monje and Raizada, 2011). Plant associated microbes have the potential to be used as biocontrol, the use of living organisms to suppress crop disease (Eilenberg, 2006) through various mechanisms including the production of antibiotics (Compant et al., 2005). Diverse classes of antimicrobial secondary metabolites of microbial origin have been reported (Mousa and Raizada, 2013), including polyketides, non-ribosomal peptides, terpenoids, heterocylic nitrogenous compounds, volatile compounds, bacteriocins as well as lytic enzymes. Polyketides and non-ribosomal peptides constitute the majority of microbial derived natural products (Cane, 1997). Interestingly, the tremendous structural diversity of antimicrobial secondary metabolites originated via limited metabolic pathways utilizing few primary metabolites as precursors (Keller et al., 2005). Underlying the diversification of antimicrobial metabolites must have been a corresponding genetic diversification of ancestral genes driven by co-evolutionary pressures (Vining, 1992).
The revolution in genomics, genome mining tools and bioinformatics offers a new opportunity to connect biochemical diversity to the underlying genetic diversity and to analyze the evolutionary events leading to biodiversity (Zotchev et al., 2012; Scheffler et al., 2013; Deane and Mitchell, 2014).
The scope of this review is to describe the biodiversity of biosynthetic coding sequences (CDS) of plant-associated microbes (bacteria and fungi) that encode selected examples of antimicrobial secondary metabolites and lytic enzymes. For each example, the target pathogen(s) and mode of action are described where known, in order to highlight the diversity of biochemical targets. Out of necessity, the review focuses on compounds for which in depth molecular analysis has been conducted. We review data pertaining to the underlying molecular diversity and highlight comparative genomic data of the orthologous genes. The review concludes with a discussion of common themes and gaps in the literature, and discusses the role of evolution in the diversification of biosynthetic gene clusters including horizontal gene transfer (HGT).
Biosynthetic Genes Encode Diverse Chemical Classes of Anti-Microbial Compounds
The diversity of compounds described in this review, the underlying genes, microbes, and pathogenic targets are summarized (Tables 1, 2).
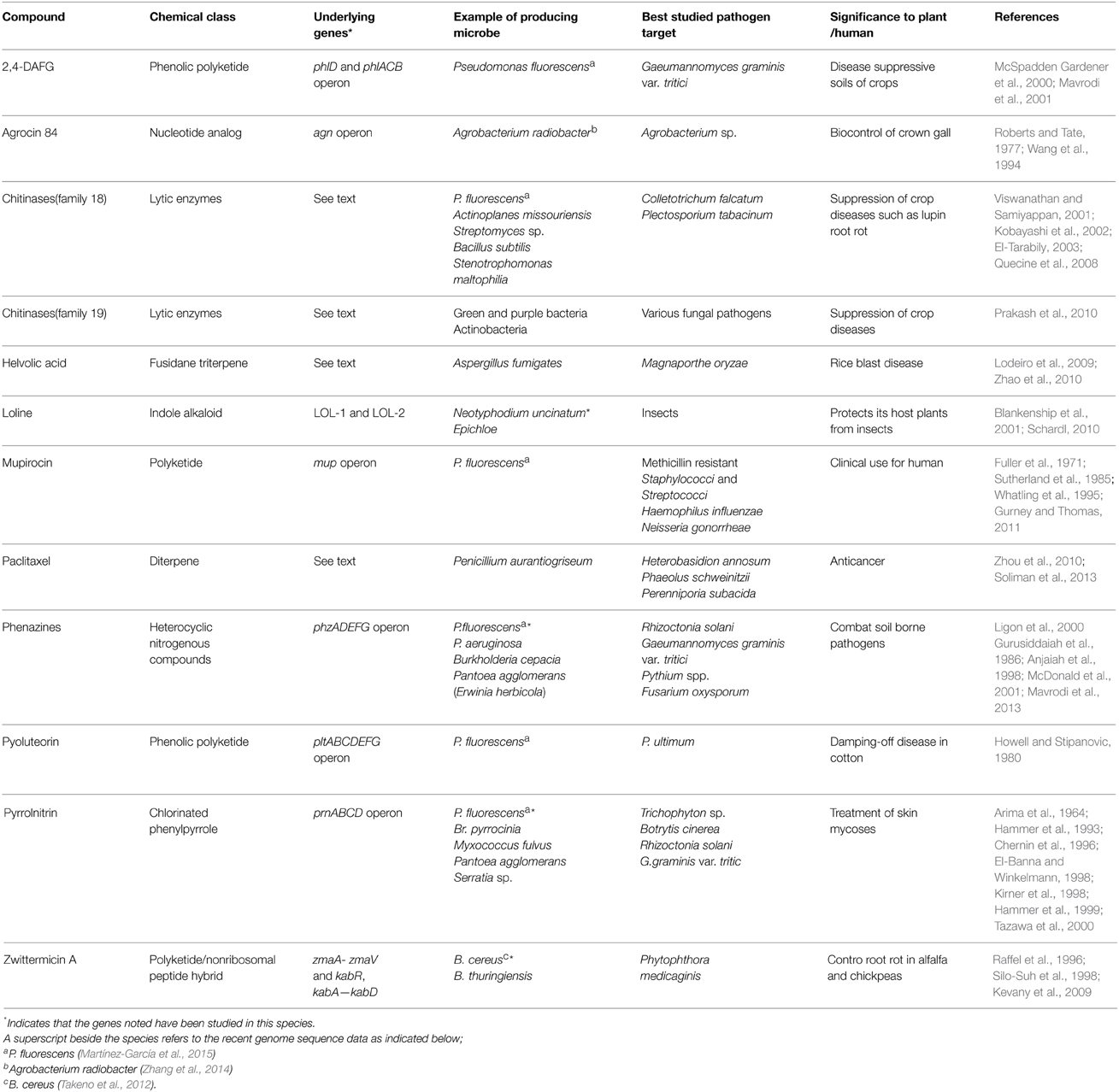
Table 1. Summary of anti-microbial compounds belonging to genetic pathways that show evidence of horizontal gene transfer (HGT).
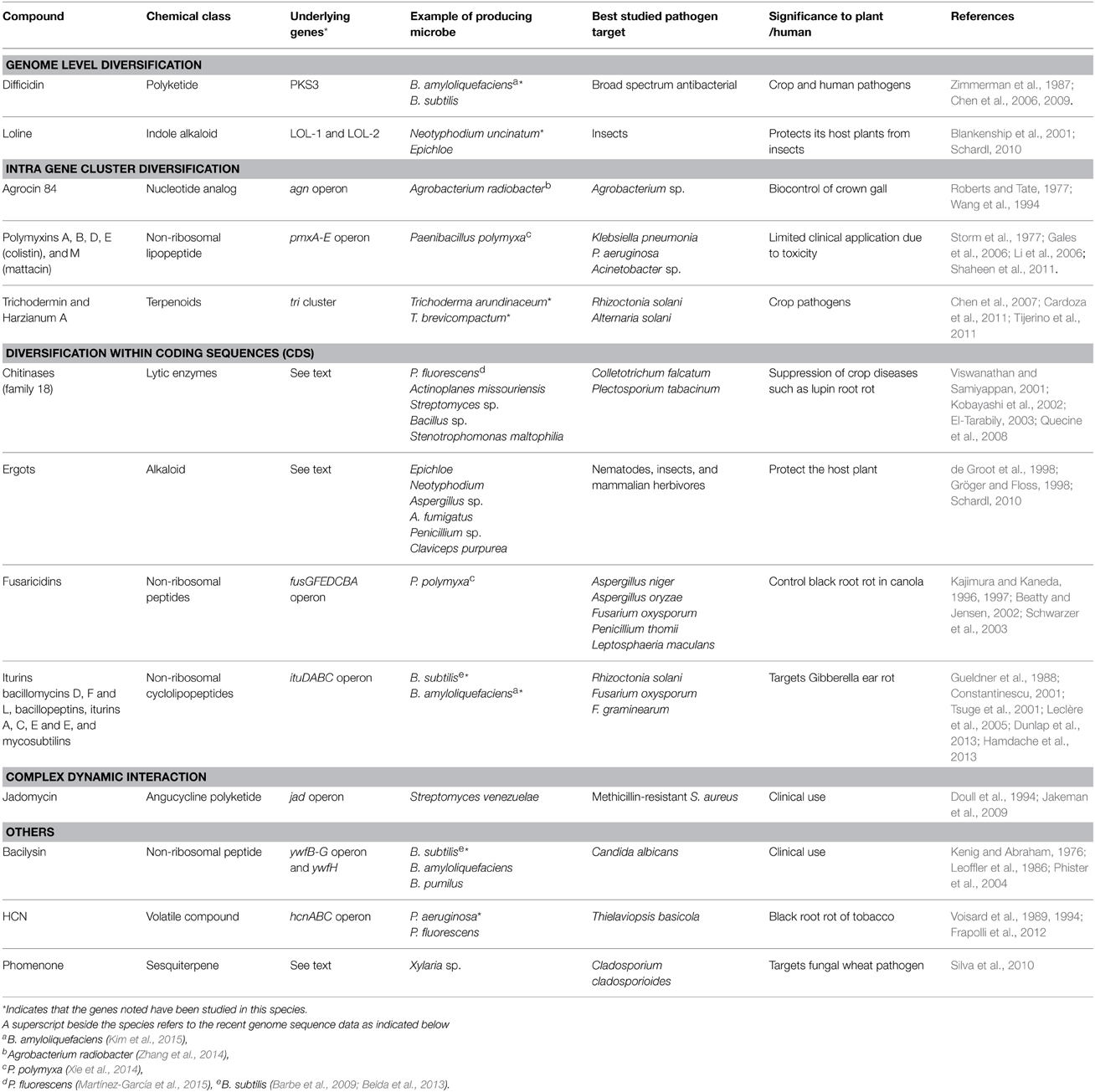
Table 2. Summary of anti-microbial compounds belonging to genetic pathways, grouped by different evolutionary levels of diversification.
Polyketides
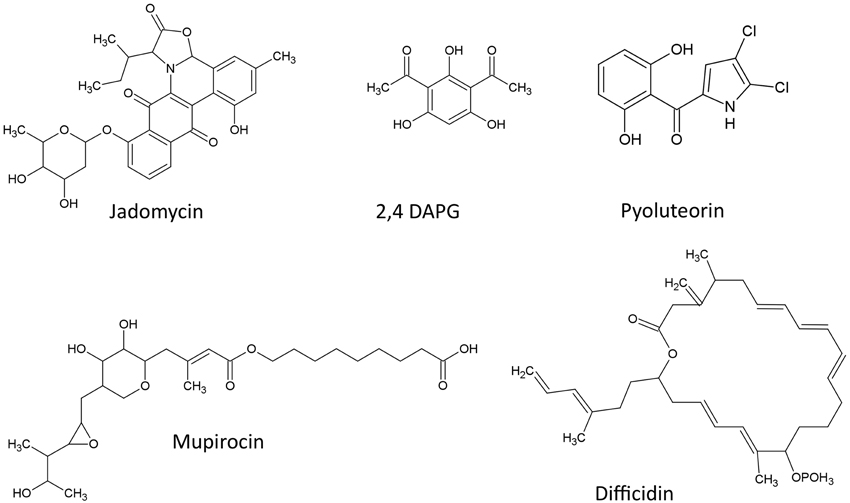
The structures of Polyketides described in this review are shown (Figure 1)
2,4-Diacetylphloroglucinol
2,4-diacetylphloroglucinol (2,4-DAPG) is a well-studied fluorescent polyketide metabolite produced by many strains of fluorescent Pseudomonas spp. that contributes to disease-suppressive soils of crops (McSpadden Gardener et al., 2000; Mavrodi et al., 2001). 2,4-DAPG is synthesized by the condensation of three molecules of acetyl coenzyme A and one molecule of malonyl coenzyme A to produce the precursor monoacetylphloroglucinol (MAPG) (Shanahan et al., 1992). In P. fluorescens strain Q2-87, four coding sequences (CDS) within the phl operon are responsible for biosynthesis of 2,4-DAPG: a single CDS (phlD) encoding a type III polyketide synthase is responsible for the production of phloroglucinol from the condensation of three acetyl-CoAs, and then three CDS (phlACB) encoding acetyltransferases are sufficient to convert phloroglucinol to 2,4-DAPG via MAPG (Bangera and Thomashow, 1999; Yang and Cao, 2012). It was suggested that the peptides encoded by phlACB may exist as a multi-enzyme complex (Bangera and Thomashow, 1999). phlD has been the subject of interest, because it has homology to chalcone and stilbene synthases from plants, which suggests horizontal gene transfer (HGT) between plants and their rhizosphere microbial populations (Bangera and Thomashow, 1999). Whereas, phlACB coding sequences are highly conserved between eubacteria and archaebacteria (Picard et al., 2000), a considerable degree of polymorphism was reported for phlD (Mavrodi et al., 2001). phlA transcription is negatively regulated by the product of phlF (Delany et al., 2000) which also appears to mediate repression by fusaric acid (Delany et al., 2000), a metabolite of pathogenic fungi of plants, that has previously been implicated in repression of biosynthesis of the anti-fungal compound, phenazine (see above) (van Rij et al., 2005). These observations demonstrate the ongoing arms race between plants, their fungal pathogens and associated anti-fungal antagonists, leading to gene diversification.
Mupirocin
The polyketide mupirocin or pseudomonic acid is one of the major antibacterial metabolites produced by Pseudomonas fluorescens (Fuller et al., 1971) and is widely used as a clinical antibiotic (Gurney and Thomas, 2011). Mupirocin can inhibit the growth of methicillin resistant Staphylococci, Streptococci, Haemophilus influenza, and Neisseria gonorrheae (Sutherland et al., 1985). In terms of the mode of action, mupirocin inhibits isoleucyl-tRNA synthetase, and hence prevents incorporation of isoleucine into newly synthesized proteins, thus terminating protein synthesis (Hughes and Mellows, 1980). Biochemically, mupirocin has a unique chemical structure that contains a C9 saturated fatty acid (9-hydroxynonanoic acid) linked to C17 monic acid A (a heptaketide) by an ester linkage (Whatling et al., 1995). Mupirocin is derived from acetate units incorporated into monic acid A and 9—hydroxynonanoic acid via polyketide synthesis (Whatling et al., 1995). At the molecular level, the mupirocin biosynthetic gene cluster (mup operon) in P. fluorescens is complex, and includes 6 Type I polyketide synthases that are multifunctional as well as 29 proteins of single function within a 65 kb region, which are incorporated into 6 larger coding sequences (modules mmpA-F) (El-Sayed et al., 2003; Gurney and Thomas, 2011). The gene cluster is non-standard as the CDS are not in the same order as the biosynthetic steps (El-Sayed et al., 2003; Gurney and Thomas, 2011). The acyltransferase (AT) domains of the polyketide synthases (PKS) are not present in each genetic module but are instead encoded by a separate CDS (from the mmpC module) and this classifies these PKS as in-trans AT PKSs (El-Sayed et al., 2003). With respect to gene regulation, two putative regulatory genes, mupR and mupI, were identified within the cluster that are involved in quorum sensing (QS) dependent regulation (El-Sayed et al., 2001).
An interesting feature of this system in P. fluorescens is that self-resistance to mupirocin is also encoded by a CDS (mupM) within the biosynthetic gene cluster (El-Sayed et al., 2003). mupM encodes a resistant Ile t-RNA synthetase (IleS) due to polymorphisms within the binding site of mupirocin (El-Sayed et al., 2003; Gurney and Thomas, 2011). A second resistant IleS was cloned from P. fluorescens NCIMB 10586 outside of the mup gene cluster which showed 28% similarity to the mupM product (Yanagisawa et al., 1994). Human pathogens that have high level mupirocin-resistance are associated with an additional gene that encode a novel IleS with similarity to eukaryotic counterparts; this resistance gene is associated with transposable elements and is carried on plasmids, facilitating its rapid spread (Eltringham, 1997; Gurney and Thomas, 2011).
There is also genetic evidence that the entire mup gene cluster in Pseudomonas arose by horizontal gene transfer; specifically the genes encoding tRNAVal and tRNAAsp were found upstream of the mupA promoter region leading to speculation that the mup cluster arose from homologous recombination between chromosomal tRNA genes and possibly a plasmid containing the mup cluster (El-Sayed et al., 2003). The inclusion of a resistant IleS (mupM) within the mup biosynthetic cluster might have facilitated such horizontal gene transfer, as otherwise uptake of the mupirocin gene cluster would have been immediately suicidal.
Difficidin
Difficidin is a polyketide with an interesting geometry that involves four double bonds in the Z configuration (Chen et al., 2006). Difficidin is produced by various Bacillus species such as B. subtilis and B. amyloliquefaciens FZB 42 with broad antibacterial activity against human and crop pathogens (Zimmerman et al., 1987; Chen et al., 2006, 2009). A large gene cluster (pks3) encoding difficidin (and oxydifficidin) was characterized in B. amyloliquefaciens (Chen et al., 2006). This compound is included in this review, because pks3 is adjacent to other polyketide synthesis gene clusters, pks1 and pks2, that encode bacillaene and macrolactin, respectively (Chen et al., 2006; Schneider et al., 2007). All three gene clusters share sequence homology, a similar order of CDS and are located close to another on the chromosome, leading Chen et al. (2006) to hypothesize that they emerged from homologous recombination from a common ancestral gene cluster resulting in gene duplication. This system provides insights into the diversification of polyketides.
Pyoluteorin
Pyoluteorin (PLt) is a phenolic polyketide with bactericidal, herbicidal, and fungicidal properties. Plt can suppress damping-off disease in cotton caused by the fungus, Pythium ultimum (Howell and Stipanovic, 1980). Both PLt and phenazine (see below) may act synergistically to suppress such soil-borne fungal diseases in plants, as some studies have suggested that the two biosynthetic pathway interact with one another (Ge et al., 2007; Lu et al., 2009). The biosynthesis of Plt involves condensation of proline with three acetate equivalents through chlorination and oxidation. The carbon skeleton is built up by the action of a single multienzyme complex (Nowak-Thompson et al., 1999). In Pseudomonas fluorescens Pf-5, a 24 kb segment contains the PLt biosynthetic operon (pltABCDEFG). PLt biosynthesis is catalyzed by type I polyketide synthases (pltB, pltC), an acyl-CoA dehydrogenase (pltE), an acyl-CoA synthetase (pltF), a thioesterase (pltG), and halogenases (pltA, pltD, pltM) with pltM located adjacent to the gene cluster (Nowak-Thompson et al., 1999). A significant delay in the expression of the PLt biosynthetic operon was reported in the cucumber spermosphere compared to cotton, which correlated to the timing of infection with the fungal root pathogen Pythium ultimum (Kraus and Loper, 1995). The authors suggest that such temporal differences may be responsible for differential disease suppression in diverse plant hosts.
The plt biosynthetic operon has been shown to be regulated by a LysR family transcriptional activator, encoded by pltR (Nowak-Thompson et al., 1999). Interestingly, pltR is tightly linked and transcribed divergently to the biosynthetic gene cluster (Nowak-Thompson et al., 1999). In earlier studies involving the biosynthetic operon of phenazine, its LysR transcriptional regulator gene (phzR) was also shown to be tightly linked to its corresponding biosynthesis gene cluster (Pierson et al., 1998). As both phenazine and PLt combat soil-borne fungal diseases in plants, we speculate that strong evolutionary pressures in the rhizosphere may have promoted HGT of the biosynthetic operons to new rhizosphere microbial hosts; the activator-cluster gene module would facilitate activation of the biosynthetic CDS following such gene transfer.
Jadomycin
Jadomycin is a member of angucycline antibiotics produced by Streptomyces species such as S. venezuelae. Jadomycin (jad) production is induced under stress conditions such as phage infection or heat shock (Doull et al., 1994; Jakeman et al., 2009). The jad biosynthetic gene cluster in S. venezuelae is closely related to type II polyketide synthase genes (Han et al., 1994) with a complex biosynthetic gene cluster (Zou et al., 2014). Jadomycin is of interest here because upstream of the jad operon are sets of negative regulatory genes including jadR1R2R3 and jadW123 (Yang et al., 1995; Zou et al., 2014). jadW123 encodes enzymes for the biosynthesis of gamma-butyrolactones (GBL), whereas JadR2 is a pseudoreceptor for GBL which upon its binding activates JadR1 and JadR3 that subsequently act as positive and negative transcriptional regulators of the jad biosynthetic operon, respectively (Zou et al., 2014). GBLs are becoming well known as regulators of secondary metabolism in gram positive bacteria, analogous to the related acyl homoserine lactone compounds which mediate QS in gram negative bacteria (Nodwell, 2014). QS is a method of communication between bacterial populations that activates genes based on high cell density through the signal molecule N-acyl-homoserine lactone (AHL) (Whitehead et al., 2001). Whereas, QS signaling molecules are thought to be synthesized and sensed by the same species (Nodwell, 2014), the GBL/jad system is interesting, because recent data suggests that GBL can signal across different Streptomyces species to activate different polyketide biosynthetic pathways (Nodwell, 2014; Zou et al., 2014). Biologically, it has been shown that different Streptomyces species, which are soil microbes, can live on the same grain of soil alongside a diversity of bacteria (Keller and Surette, 2006; Vetsigian et al., 2011), suggesting there may have been evolutionary selection for inter-species coordination for antibiotic production (Nodwell, 2014), resulting in enhanced genetic complexity associated with the jad locus.
Non-Ribosomal Peptides
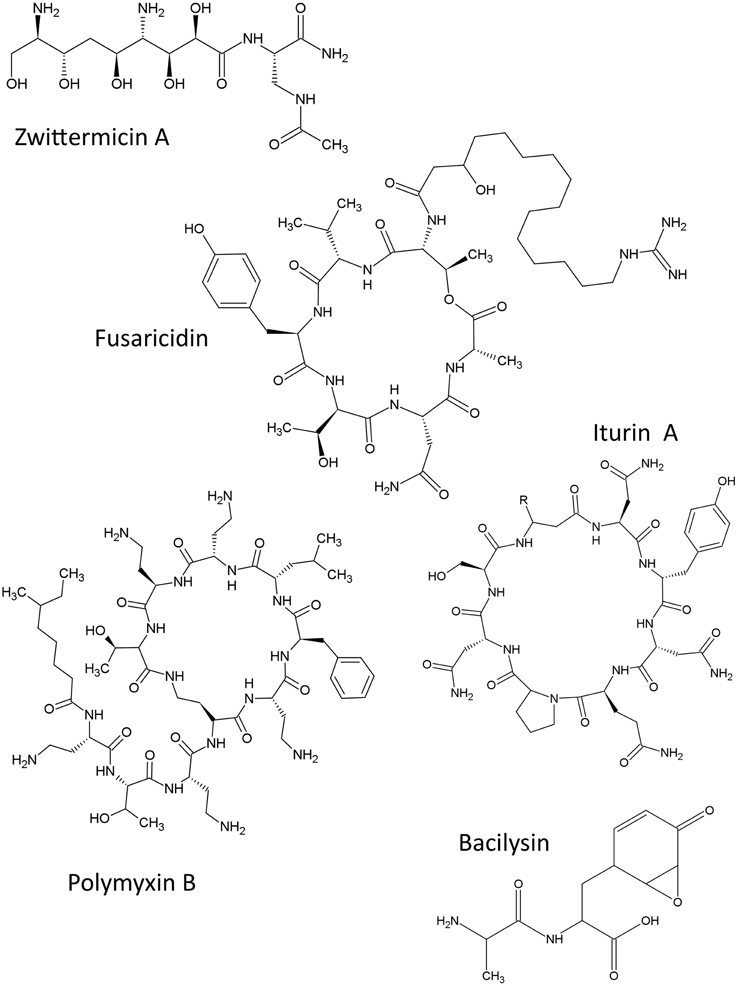
The structures of non-ribosomal peptides described in this review are shown (Figure 2).
Zwittermicin A
Zwittermicin A is a polyketide/nonribosomal peptide hybrid antibiotic produced by B. cereus and B. thuringiensis (Raffel et al., 1996) with activity against oomycetes such as Phytophthora medicaginis and some other pathogenic fungi (Silo-Suh et al., 1998). Zwittermicin A has a unique structure that includes glycolyl moieties, D amino acid, and ethanolamine in addition to the unusual terminal amide produced from the ureidoalanine (nonproteinogenic amino acid) (Kevany et al., 2009). Zwittermicin A is thought to be biosynthesized as part of a larger metabolite that is processed twice to form zwittermicin A and two other metabolites (Kevany et al., 2009). The complete biosynthetic operon encoding zwittermicin A includes 27 open reading frames (CDS, zmaA, and zmaV) that extend over 62.5 kb of the Bacillus cereus UW85 genome, in addition to five individual genes (kabR and kabA—kabD) (Kevany et al., 2009). In this study, support was gained for the hypothesis that the skeleton of zwittermicin A is catalyzed by a megasynthase enzyme involving multiple nonribosomal peptide synthetases (NRPS) and PKS; the megasynthase has multiple modules containing distinct domains that catalyze the different steps in the pathway (Emmert et al., 2004; Kevany et al., 2009). Evidence suggested that the CDS included 5 NRPS modules (Kevany et al., 2009). It is noteworthy that a similar gene cluster was characterized on a plasmid in B. cereus AH1134, suggesting that the pathway can be transferred horizontally (Kevany et al., 2009). Consistent with the mobility of this operon, an orthologous 72-kb region encoding for zwittermicin A in Bacillus thuringiensis, was shown to be flanked by putative transposase genes on both edges, suggesting that it may be a mobile element that was gained by B. cereus through horizontal gene transfer. Since zwittermicin A has been reported to enhance the activity of protein toxins that attack insects (Broderick et al., 2000), it was hypothesized that transfer of this operon into B. thuringiensis permitted the microbe to gain insecticide-promoting factors to combat insects during co-evolution (Luo et al., 2011).
Fusaricidins A–D
Fusaricidins are guanidinylated ß-hydroxy fatty acids attached to a cyclic hexapeptide including four D-amino acids (Kajimura and Kaneda, 1997; Schwarzer et al., 2003). These antibiotics are produced by Paenibacillus polymyxa strains and exhibit antifungal activity against diverse plant pathogens including, Aspergillus niger, Aspergillus oryzae, Fusarium oxysporum, and Penicillium thomii (Kajimura and Kaneda, 1996, 1997) as well as Leptosphaeria maculans, the causal agent of black root rot in canola (Beatty and Jensen, 2002). The amino acid chains of fusaricidins are linked together and modified by a non-ribosomal peptide synthetase (NRPS). The multi-domain NRPS consists of up to 15,000 amino acids and is therefore considered among the longest proteins in nature (Schwarzer et al., 2003). NRPS incorporation is not limited to the 21 standard amino acids translated by the ribosome, and this promiscuity contributes to the great structural diversity and biological activity of non-ribosomal peptides (Li and Jensen, 2008).
In P. polymyxa E68, the fusaricidin biosynthetic gene cluster (fusGFEDCBA) has been characterized in which the NRPS coding sequence, the largest CDS in the cluster, was observed to encode a six-module peptide (Choi et al., 2008; Li and Jensen, 2008; Li et al., 2013). The biosynthetic cluster includes other CDS responsible for biosynthesis of the lipid moiety but does not contain transporter genes (Li and Jensen, 2008). In P. polymyxa, a promoter for the fus operon was identified and shown to be bound by a transcriptional repressor (AbrB) which previous studies implicated as a regulator of sporulation; this is of interest since fusaricidin was observed to be synthesized during sporulation, thus coordinating the microbe's secondary metabolism with its life cycle (Li et al., 2013).
Allelic diversity is typically thought to be responsible for producing chemical diversity. However, an interesting feature of the fus cluster is that a diversity of fusaricidins, differing in their incorporated amino acids (Tyr, Val, Ile, allo-Ile, Phe), can be produced by a single allele of fusA; the underlying mechanism is that the NRPS A-domain, responsible for recognition of amino acids, has relaxed substrate specificity (Figure 3) (Han et al., 2012).

Figure 3. Diagram illustrating how a diversity of fusaricidins are produced from a single allele of fusA which encodes the non-ribosomal peptide synthase (NRPS) A-domain. (A) Most enzymes have stringent substrate specificity. (B) By contrast, the NRPS A-domain can recognize and incorporate different amino acids to create diverse fusaricidins, and hence it is an example of an enzyme with relaxed substrate specificity (Han et al., 2012).
Polymyxins
Polymyxins are a family of non-ribosomal lipopeptide antibiotics composed of ten amino acids, a polycationic heptapeptide ring and a fatty acid derivative at the N terminus (Storm et al., 1977). They are produced by Gram positive bacteria and target Gram negative species, by altering the structure of the cell membrane. The polymyxin family includes polymyxins A, B, D, E (colistin), and M (mattacin) (Shaheen et al., 2011). Polymyxin B exhibits potent antibacterial activity against Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. (Gales et al., 2006). However, polymyxins exhibit a remarkable degree of neurotoxicity and nephrotoxicity which limit their clinical use (Li et al., 2006).
In Paenibacillus polymyxa PKB1 (the same strain that controls plant fungi by producing fusaricidins, see above), a 40.8 kb polymyxin biosynthetic gene cluster was shown to encode five coding sequences, pmxA-E. Three CDS (pmxA, B, E) encode subunits of NRPS, each responsible for the modular incorporation of amino acids, while two genes (pmxC, D) encode a permease belonging to the ABC-type transporter family (Shaheen et al., 2011). In both P. polymyxa PKB1 and P. polymyxa E681, the arrangement of the NRPS coding sequences in the pmx cluster does not match the amino acid sequence in the produced polymyxin, which is unusual for NRPS-encoded peptides (Choi et al., 2009; Shaheen et al., 2011).
With respect to the diversity of polymyxins, polymyxins differ in the amino acid composition of residues 3, 6, and 7, in the D vs. L stereochemistry of the incorporated amino acids, as well as in the lipid moiety (Choi et al., 2009; Shaheen et al., 2011). In P. polymyxa SC2 and P. polymyxa PKB1, an allelic variant was uncovered within NRPS domain 3 using bioinformatic analysis of the genome which correlated with incorporation of the D rather than L form of 2,4-diaminobutyrate in amino acid position 3, explaining the mechanism for the production of two subtypes of polymyxin B (Shaheen et al., 2011). With respect to the diversity of residues 6 and 7, P. polymyxa E681 and P. polymyxa PKB1 produce polymyxins that differ in these amino acids, producing polymyxin A and B, respectively (Shaheen et al., 2011). Bioinformatic analysis revealed that the DNA sequences of the pmx gene clusters were 92% conserved at the nucleotide level, but differed considerably in the domains corresponding to modules 6 and 7 (Shaheen et al., 2011). These two sets of observations led the authors to suggest that the diversity of polymyxins arises from mixing and matching of alleles of the NRPS modular domains, hence combinatorial chemistry, rather than relaxed substrate specificity as seen in other secondary metabolites such as fusaricidins (see above).
Another interesting feature of the pmx gene clusters is that the polymyxin transporters might also transport fusaricidin, since the fus biosynthetic cluster lacks any transporter genes (see above), and as both antibiotics are cationic lipopeptides (Shaheen et al., 2011). The authors found support for this hypothesis, as deletion mutations in pmxC and D genes also reduced the antifungal activity of fusaricidin against Leptosphaeria maculans although the two biosynthetic gene clusters are not linked. It is worth noting that there is no evidence yet of genes responsible for lipidation of the peptide residue in the characterized polymyxin clusters, suggesting that this function might be encoded elsewhere in the genome (Shaheen et al., 2011).
Iturins
Iturins are a family of non-ribosomal cyclolipopeptides consisting of seven α-amino acid residues and one ß-amino acid, the latter noted as a unique feature compared to other lipopeptide antibiotics (Constantinescu, 2001; Leclère et al., 2005; Hamdache et al., 2013). The iturin family includes compounds such as bacillomycins D, F and L, bacillopeptins, iturins A, C, E and E, and mycosubtilins (Hamdache et al., 2013). Iturins are produced by different strains of B. subtilis and B. amyloliquefaciens, and exhibit potent antifungal activity against major phytopathogens including R. solani, Fusarium oxysporum, and F. graminearum, the latter responsible for Fusarium head blight in wheat (Gueldner et al., 1988; Constantinescu, 2001; Tsuge et al., 2001; Dunlap et al., 2013). The mechanism of action involves disruption of the target fungal plasma membrane (Thimon et al., 1995). In both B. subtilis RB14 and B. amyloliquefaciens AS43.3, the iturin A biosynthetic operons were shown to contain four coding sequences (ituDABC) coding for: a putative malonyl coenzyme A transacylase, a protein with three functions (fatty acid synthetase, amino acid transferase, and peptide synthetase), and two peptide synthetases, respectively (Tsuge et al., 2001; Dunlap et al., 2013).
Regarding diversification within the chemical family, iturin A from B. subtilis RB14 has a similar structure as mycosubtilin that is produced by B. subtilis ATCC 6633 but with inverted amino acids at the 6th and 7th positions (Tsuge et al., 2001). By comparative analysis of orthologous CDS between these two strains (ituC and mycC, respectively), it was suggested that the NRPS amino acid adenylation domain may have been intragenically swapped during evolution, which would also imply a HGT event (Tsuge et al., 2001). Comparative genome analysis between at least three sequenced itu clusters may reveal further information concerning the diversification of the iturin family (Tsuge et al., 2001; Blom et al., 2012; Dunlap et al., 2013).
Bacilysin
Bacilysin is a non-ribosomally produced dipeptide composed of an L-alanine residue at the N terminus and a non-proteinogenic amino acid, L-anticapsin, at the C terminus (Walker and Abraham, 1970; Stein, 2005). Compared to the more elaborate non-ribosomal peptides noted above, bacilysin is noteworthy because it is amongst the simplest peptides in nature, adding to the structural diversity of observed non-ribosomal peptides. Bacilysin is produced by Bacillus species such as B. pumilus, B. amyloliquefaciens, and B. subtilis (Leoffler et al., 1986; Phister et al., 2004) and shown to have antimicrobial activity against various bacteria and fungi such as Candida albicans (Kenig and Abraham, 1976). Mechanistically, bacilysin is a prodrug that is activated by the action of a peptidase enzyme that releases the active moiety, anticapsin (Rajavel et al., 2009). Anticapsin inhibits bacterial peptidoglycan or fungal protein biosynthesis through blockage of glucosamine synthetase, resulting in cell lysis (Kenig et al., 1976). Biosynthesis of bacilysin originates from the prephenate aromatic amino acid pathway (Hilton et al., 1988; Parker and Walsh, 2012).
In B. subtilis the biosynthesis of bacilysin is encoded by the operon, bacABCDE (ywfB-G), in addition to a monocistronic gene (ywfH) (Inaoka et al., 2003). bacABC is likely responsible for the biosynthesis of anticapsin while bacDE (ywfEF) encodes a ligase and an efflux transporter protein for self protection, respectively (Steinborn et al., 2005; Rajavel et al., 2009). The bacilysin biosynthetic operon is positively regulated by QS pheromones, in particular PhrC (Yazgan et al., 2001; Köroğlu et al., 2011) and negatively regulated by ScoC, a transition state regulator (Inaoka et al., 2009). The transition state in bacteria is a period of decision making.
Terpenoids
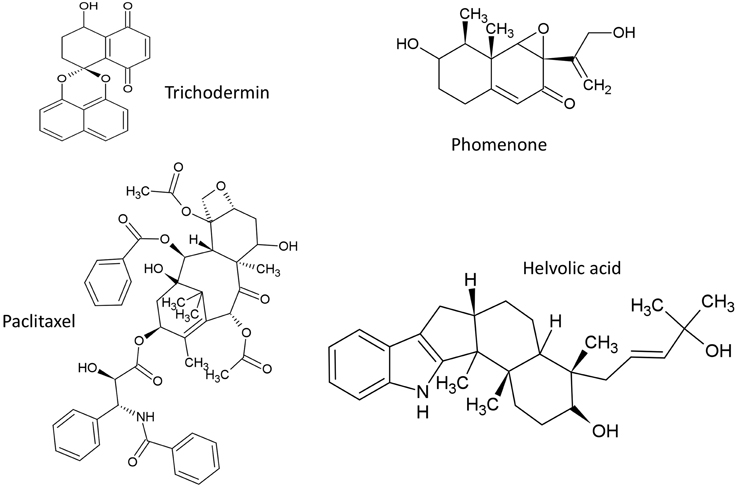
The structures of terpenoids described in this review are summarized (Figure 4).
Trichodermin and Harzianum A
Trichothecene mycotoxins are produced by some fungal genera such as deoxynivalenol (DON) from Fusarium, and harzianum and trichodermin from Trichoderma arundinaceum and T. brevicompactum, respectively (Cardoza et al., 2011). Trichodermin was reported to have antifungal activity against the fungal pathogens Rhizoctonia solani and Alternaria solani (Chen et al., 2007) as well as other fungal genera (Tijerino et al., 2011). Trichodermin inhibits protein synthesis in eukaryotes by inhibiting peptidyl transferase that catalyzes translational elongation and/or termination (Wei et al., 1974) and by inhibiting peptide-bond formation at the initiation stage of translation (Carter et al., 1976).
Comparative analysis has been conducted on the CDS responsible for trichothecene biosynthesis in Fusarium and Trichoderma. In Fusarium, trichothecenes are encoded by a gene cluster called the TRI cluster; this cluster also encodes regulatory and transport proteins (Proctor et al., 2009). In Trichoderma, an orthologous TRI cluster was discovered in which 7 CDS were conserved with Fusarium, but the two clusters showed interesting evolutionary divergence (Cardoza et al., 2011) which may be informative for understanding the genetics underlying other anti-fungal metabolites. In Fusarium, the TRI cluster includes tri5 that encodes trichodiene synthase, the first committed step in trichothecene biosynthesis, which catalyzes the cyclization of farnesyl pyrophosphate to form trichodiene (Hohn and Beremand, 1989). In Fusarium, tri5 is located within the TRI cluster, but surprisingly it is not associated with the orthologous cluster in Trichoderma. Three additional CDS responsible for trichothecene biosynthesis in Fusarium (tri7, tri8, tri13) are missing from the Trichoderma cluster, along with an CDS of unknown function (tri9) (Cardoza et al., 2011). Interestingly, two of the apparently conserved biosynthetic CDS (tri4 and tri11, based on sequence homology) were demonstrated to have diverged functionally between Trichoderma and Fusarium based on heterologous expression analysis: in Trichoderma, tri4 catalyzes three out of four oxygenation reactions carried out by its corresponding Fusarium ortholog; tri11 catalyzes distinctive hydroxylation reactions in Fusarium (C-15) and Trichoderma (C-4). Finally, amongst the CDS which are conserved between Fusarium and Trichoderma, head-to-tail vs. head-to-head rearrangements are observed (e.g., tri3, tri4) (Cardoza et al., 2011). These results demonstrate multiple evolutionary events (rearrangement, functional diversification, gene loss, gene gain) within one biosynthetic gene cluster (Figure 5).
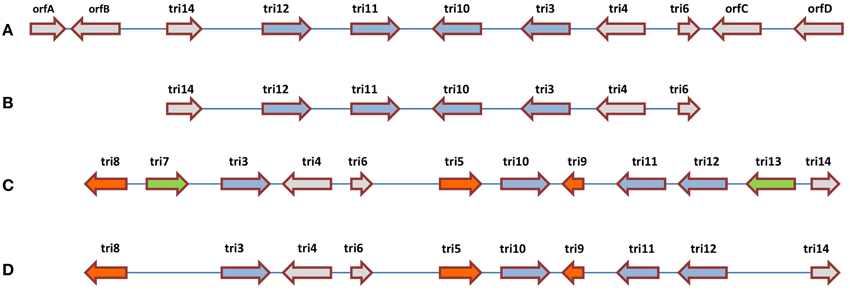
Figure 5. Comparative analysis of the trichothecene biosynthetic gene clusters in (A) Trichoderma arundinaceum, (B) T. brevicompactum, (C) Fusarium sporotrichioides, and (D) F. graminearum. The illustration suggests that the ancestral gene cluster underwent multiple evolutionary events including re-arrangements (blue arrows), gene gain or loss within the same genus (green arrows) and gene gain or loss between genera (orange and green arrows) (adapted from Cardoza et al., 2011).
Phomenone
Phomenone is a sesquiterpene synthesized by various fungi including Xylaria sp., an endophytic fungus isolated from Piper aduncum, and reported to have antifungal activity against the pathogen Cladosporium cladosporioides (Silva et al., 2010). Phomenone is structurally similar to the PR toxin metabolite of Penicillium roqueforti which functions by inhibiting RNA polymerase and thus inhibits protein synthesis at the initiation and elongation steps (Moule et al., 1976). A biosynthetic precursor for phomenone A is aristolochene (Proctor and Hohn, 1993). In P. roqueforti NRRL 849, a gene required for aristolochene (aril) biosynthesis was characterized and shown to encode a sesquiterpene cyclase named aristolochene synthase (AS) (Proctor and Hohn, 1993). Expression of aril occurs in stationary phase cultures and is regulated transcriptionally (Proctor and Hohn, 1993).
Paclitaxel (Taxol)
The diterpene paclitaxel (Taxol) is reported to be produced by at least 20 diverse fungal endophyte genera inhabiting various plant species (Zhou et al., 2010). Taxol was reported to be produced by some fungal endophytes that inhabit conifer wood and its ecological function was suggested to be a fungicide against host pathogens (Soliman et al., 2013). Taxol acts by stabilizing microtubules and inhibiting spindle function leading to disruptions in normal cell division (Horwitz, 1994). However, Taxol was originally purified from Taxus trees (Wani et al., 1971) and shown to be encoded by plant nuclear genes, apparently redundantly. As the number of plant genera that produce Taxol is very few, it is interesting to speculate whether its biosynthetic genes may have been transferred horizontally from fungi to plants.
The Taxol biosynthetic pathway in plants requires 19 enzymatic steps. The first committed step in biosynthesis of plant Taxol is cyclization of GGDP to taxa-(4,5),(11,12)-diene catalyzed by taxadiene synthase (TS) (Hezari et al., 1995). Thirteen plant Taxol biosynthetic genes from Taxus were used in BLASTP searches to identify potential homologs in Penicillium aurantiogriseum NRRL 62431 (Yang et al., 2014). Seven putative homologous genes were identified though the homology scores were as low as 19%; these genes were claimed to encode: phenylalanine aminomutase (PAM), geranylgeranyl diphosphate synthase (GGPPS), taxane 5α-hydroxylase (T5OH), taxane 13α-hydroxylase (T13OH), taxane 7β-hydroxylase (T7OH), taxane2α-hydroxylase (T2OH) and taxane 10β-hydroxylase (T10OH). Another gene encoding an AT (PAU_P11263) was identified by using BLASTP against the GenBank database. However, no homologs were identified to plant TS; the authors claimed that the fungus might catalyze taxadiene synthesis by a unique enzymatic system (Yang et al., 2014). Position-Specific Iterative BLAST showed one gene from the bacterial genus Mycobacterium with potential similarity to plant TS suggesting lateral gene transfer from plants to mycobacteria (Yang et al., 2014).
In a parallel study to isolate fungal Taxol biosynthetic genes, a different approach was taken where PCR primers designed from the plant genes that encode Taxol were used as a primary screen against fungi (Xiong et al., 2013). The study identified putative homologs of fungal TS as well as BAPT (which encodes the critical C-13 phenylpropanoid side-chain CoA acyltransferase) with ~40% sequence identities to their plant counterparts. Despite this progress, other reports remain skeptical that fungi actually encode Taxol (Heinig et al., 2013).
Recent studies have demonstrated complex three-way interactions in Taxol biosynthesis between a Taxol-producing fungal endophyte, other endophytes and the host plant. Host endophytic fungi appear to elicit plant TS transcription or transcript accumulation. Specifically, TS transcript and the corresponding protein were reduced upon treating both young plantlets and old Taxus wood with fungicide (Soliman et al., 2013). In a parallel study, co-culture of the Taxol-producing endophyte Paraconiothyrium SSM001 with two presumptive fungal endophytes of the same yew tree host elicited paclitaxel accumulation from the endophyte, suggesting inter-species interactions between endophytes inhabiting the same host niche (Soliman and Raizada, 2013).
Helvolic Acid
Helvolic acid is a fusidane triterpene produced by Aspergillus fumigatus (Lodeiro et al., 2009) and the yeast, Pichia guilliermondii Ppf9 (Zhao et al., 2010). Helvolic acid was reported to inhibit the spore germination of Magnaporthe oryzae, the causal agent of rice blast disease (Zhao et al., 2010). The biosynthetic genes for helvolic acid are clustered as nine genes coding for protostadienol synthase which catalyzes the precursor (17Z)-protosta-17(20),24-dien-3-ol, along with genes that encode squalene-hopene cyclase, four cytochrome P450 monooxygenases, short chain dehydrogenase, two transferases and 3-ketosteroid 1-dehydrogenase (Lodeiro et al., 2009). The authors reported that the P450 monooxygenases from different fungi shared substantial sequence identity across recent evolution, while the transferases duplicated and diversified into paralogous gene families (Lodeiro et al., 2009). This observation suggests that even within a single gene cluster, there may be different selection pressures on adjacent genes belonging to the same biosynthetic pathway. Interestingly, the helvolic acid biosynthetic gene cluster in A. fumigates is located in the sub-telomere chromosome region (Lodeiro et al., 2009) which is associated with high rates of evolutionary recombination and diversification. However, the gene cluster lacks introns which is a trait sometimes associated with subtelomeric regions, but this observation might also be evidence of HGT from bacteria (Lodeiro et al., 2009).
Alkaloids
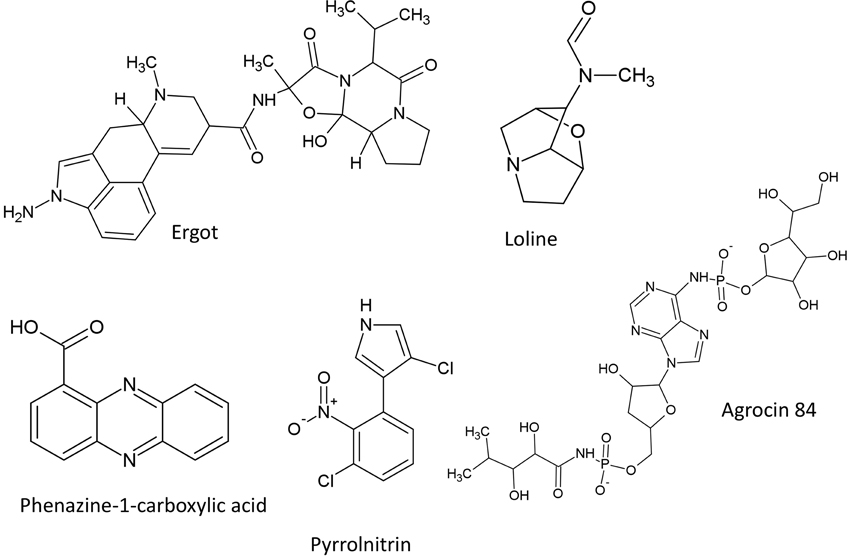
The structures of alkaloids described in this review are summarized (Figure 6).
Ergot
Ergot alkaloids are produced from the sexual Epichloe fungi and their asexual derivatives Neotyphodium within the Clavicipitaceae family which inhabit Pooideae grasses (Schardl, 2010). Ergot alkaloids can interact with receptors of the central nervous system and exhibit toxic effect on nematodes, insects, and mammalian herbivores including livestock which graze these grasses (de Groot et al., 1998; Gröger and Floss, 1998). In Europe in the Middle Ages, consumption of ergot-infected grain or grasses caused convulsions, paranoia and hallucinations in livestock and humans, known as St. Anthony's Fire (Dotz, 1980). The diverse ergot alkaloids share a tetracyclic ergoline backbone derived from tryptophan and dimethylallyl diphosphate (Flieger et al., 1997). Gene clusters for ergot alkaloid biosynthesis have been identified in various Ascomycete species belonging to Aspergillus, Penicillium, and Claviceps. Seven genes encode the ergoline scaffold including dimethylallyltryptophan synthase (DMATS) which catalyzes the first committed step. DMATS is responsible for the prenylation of L-tryptophan with dimethylallylpyrophosphate (DMAPP) to produce 4-dimethylallyltryptophan (4-DMAT) (Heinstein et al., 1971). Ergots have diversified into three classes, caused by diverse substituents attached to the carboxyl group of the tetracyclic ergoline backbone, in particular the presence of an amide group (creating ergoamides), a peptide-like amide moiety (creating ergopeptines) or the absence of these moieties (creating clavine alkaloids) (Wallwey and Li, 2011). These structural modifications are responsible for the differential physiological and pharmacological effects of the ergot family, that include treatment of postpartem hemorrhage, leukemia, and Parkinson's disease. The genetic basis for ergot diversification into these 3 major classes is associated with the presence or absence of nonribosomal peptide synthases (NRPS) which catalyze the biosynthesis of the peptide moieties on the ergoline backbone (Wallwey and Li, 2011). For example, four NRPS genes are present in Claviceps purpurea (which encodes ergopeptines) but absent in Aspergillus fumigatus (which produces clavine alkaloids). Inactivation of these genes suggests that two of the NRPS genes (lpsA and lpsB) are also responsible for synthesis of the ergoamides (Haarmann et al., 2008). Interestingly, further diversification of the peptide moiety within C. purpurea has been reported to be caused by fine-scale allelic diversification of the NRPS genes (Haarmann et al., 2005). There is additional evidence to suggest that diversification of the ergot alkaloid gene clusters is associated with DNA transposons and retroelements, which were observed in the cluster encoding ergovaline, an ergot alkaloid from Epichloe festucae associated with livestock toxicity (Fleetwood et al., 2007). As an interesting note, the genes encoding ergovaline were highly expressed only during biotrophic growth of the fungus within the host grass plant not when the mycelia were cultured in vitro, suggesting that the host might have a regulatory role in the expression of the fungal gene cluster (Fleetwood et al., 2007).
Loline Alkaloid
Loline is an indole alkaloid produced by Neotyphodium uncinatum fungus, the asexual mutualistic derivative of Epichloe, which is known to protect its host plants from insects (Blankenship et al., 2001; Schardl, 2010). The loline biosynthetic pathway was suggested to involve proline and homoserine (Spiering et al., 2005). In N. uncinatum, two homologous gene clusters encoding loline were identified, named LOL-1 and LOL-2 (Spiering et al., 2005). The cluster LOL-1 involves nine genes-(lolF-1, lolC-1, lolD-1, lolO-1, lolA-1, lolU-1, lolP-1, lolT-1, lolE-1) within a 25-kb chromosomal segment, while the LOL-2 cluster contains the same homologs (except for lolF) ordered and oriented the same as in LOL-1. This evidence suggests that the loline clusters may represent a recent segmental duplication event (Spiering et al., 2005).
An interesting ecological situation exists in grasses infected with Epichloe fungi (sexual form of Neotyphodium): the fungus reduces the ability of these plants to propagate sexually (they choke the inflorescences), which, without compensatory mechanisms, would prevent vertical transmission of the fungus (Zhang et al., 2010). However, to compensate, the fungal stromata attract fly vectors which transmit the fungal spores to other plants, permitting horizontal transfer of the fungus. Loline accumulates in young tissues of the grasses, providing insect protection to these young hosts; however if loline was also to accumulate in the grass inflorescences, it would kill the fly vector of the fungus. Upon further investigation, this apparent paradox was resolved: in these grass inflorescences, transcription of the loline biosynthesis genes was dramatically downregulated compared to plants with healthy inflorescences (infected with the symbiotic asexual Neotyphodium), permitting the fly vectors to survive (Zhang et al., 2010). These observations suggest strong selection pressure to evolve the regulatory elements of these genes.
Heterocyclic Nitrogenous Compounds
The structures of heterocyclic nitrogenous compounds described in this review are summarized (Figure 6).
Phenazines
Phenazines are a group of naturally occurring heterocyclic nitrogenous antibiotics produced exclusively by bacteria and widely reported in fluorescent Pseudomonas (Mavrodi et al., 2006, 2013). Phenazines are potent antifungal compounds that can combat soil borne pathogens (Ligon et al., 2000) such as Rhizoctonia solani, Gaeumannomyces graminis var. tritici, Pythium spp. (Gurusiddaiah et al., 1986) and Fusarium oxysporum (Anjaiah et al., 1998). Mechanisms of action include: (1) accumulation of toxic molecules such as hydrogen peroxide and superoxide due to the redox potential of phenazine (Hassan and Fridovich, 1980; Hassett et al., 1995); and (2) elicitation of induced host resistance (Audenaert et al., 2002). Ecologically, the evidence suggests that the plant rhizosphere promotes phenazine-producing bacteria to combat pathogens (Mazzola et al., 1992; Mavrodi et al., 2013).
Phenazine is derived from the shikimic acid pathway, with amino-2-deoxyisochorismic acid (ADIC) as the branchpoint to phenazine (McDonald et al., 2001). ADIC is then converted to trans-2, 3-dihydro-3-hydroxy anthranilic acid which undergoes dimerization to form phenazine-1-carboxylic acid, the first derivative of the phenazines (McDonald et al., 2001). Phenazine biosynthesis in Pseudomonas fluorescens is encoded by a single or duplicated core of five CDS, phzADEFG, that encode ketosteroid isomerase, isochorismatase, anthranilate synthase, trans-2,3-dihydro-3-hydroxyanthranilate isomerase, and pyridoxamine oxidase respectively (Mavrodi et al., 2013). In Pseudomonas, the core may include other CDS such as phzB which was duplicated from phzA, and phzC which encodes 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase that is responsible for diverting carbon from the shikimate pathway to phenazine (Pierson and Pierson, 2010).
Comparisons between Pseudomomas species and other genera have revealed conservation yet diversity of the core phenazine biosynthetic CDS. For example, the phenazine biosynthesis operon in Burkholderia cepacia maintains the five core enzymes observed in Pseudomonas as reviewed (Mavrodi et al., 2006). However, there is evidence to suggest that these coding sequences spread to enteric bacteria and Burkholderia species via horizontal gene transfer, because these genes can be observed in plasmids and transposons (Mavrodi et al., 2010). For example, in Erwinia herbicola, a biosynthetic cluster of 16 CDS (ehp) was isolated from a plasmid, of which 15 coded for D-alanyl griseoluteic acid (AGA) while ehpR was observed to encode for resistance to AGA (Giddens et al., 2002). Other differences in the core have also been observed between Pseudomonas species and others; for example in both Burkholderia cepacia and Erwinia herbicola, phzA is not duplicated (Mavrodi et al., 2006).
Structural diversity of phenazines in different species is achieved by specific genes that may be located within the cluster or elsewhere in the genome. For example, in P. chlororaphi, the phzH gene is located downstream of the phenazine operon, where it encodes an aminotransferase responsible for converting phenazine-1-carboxylic acid (PCA) to phenazine-1-carboxamide, the characteristic green pigment of P. chlororaphis (Chin-A-Woeng et al., 1998). In P. aureofaciens, phzO was identified as the gene that encodes an aromatic monooxygenase, responsible for catalyzing the hydroxylation of PCA to form the broad spectrum antifungal compound, 2-OH-PCA (Delaney et al., 2001). In P. aeruginosa (PAO1), two diversification genes were discovered: phzM was shown to be involved in the synthesis of pyocyanin while phzS gene encodes a monooxygenase that catalyzes the production of 1-hydroxy phenazine (Mavrodi et al., 2001).
Pyrrolnitrin
Pyrrolnitrin is a chlorinated phenylpyrrole antibiotic purified initially from Burkholderia pyrrocinia (Arima et al., 1964) then subsequently from other species including pseudomonads, Myxococcus fulvus, Enterobacter agglomerans, and Serratia sp (Chernin et al., 1996; Kirner et al., 1998; Hammer et al., 1999). Pyrrolnitrin was initially used for treatment of skin mycoses caused by Trichophyton fungus, then was developed as an effective fungicide for crops against Botrytis cinerea (Hammer et al., 1993), Rhizoctonia solani (El-Banna and Winkelmann, 1998) and Gaeumannomyces graminis var. tritici (Tazawa et al., 2000). In P. fluorescens, the pyrrolnitrin biosynthetic operon consists of four coding sequences (prnABCD) coding for tryptophan halogenase (prnA), a decarboxylase (prnB), monodechloroaminopyrrolnitrin halogenase (prnC), and an oxidase (prnD) (Hammer et al., 1997; Kirner et al., 1998). Comparative analysis indicates that the pyrrolnitrin biosynthetic operon is differentially conserved between divergent species with 59% similarity among diverse bacterial strains such as Pseudomonas, Myxococcus fulvus, and Burkholderia cepacia, with lower similarity shown for prnA in M. fulvus (45%) (Hammer et al., 1999). Furthermore, RFLP-based polymorphisms within a 786 bp prnD fragment suggested that there may have been lateral gene transfer of the prn operon from Pseudomonas to Burkholderia pyrrocinia (Souza and Raaijmakers, 2003). Consistent with such mobility, transposase-encoding genes surrounding the prn biosynthetic operon were observed in Burkholderia pseudomallei (Costa et al., 2009).
Volatile Compounds
In this section, only the most well studied volatile compound, hydrogen cyanide, is discussed.
Hydrogen Cyanide (HCN)
Hydrogen cyanide (HCN) is a volatile secondary metabolite produced by P. aeruginosa, and diverse rhizosophere fluorescent pseudomonads, where they exhibit biocontrol activity against pathogenic fungi such as Thielaviopsis basicola, the fungal causal agent of black root rot of tobacco (Voisard et al., 1989, 1994; Frapolli et al., 2012). Mechanistically, HCN functions by inhibiting important metalloenzymes such as cytochrome c oxidase (Blumer and Haas, 2000) and/or by complexing metals in the soil (Brandl et al., 2008). HCN is biosynthesized from glycine (Castric, 1977) in an oxidative reaction catalyzed by HCN synthase, a membrane-bound flavoenzyme (Castric, 1994; Blumer and Haas, 2000). The biosynthesis of HCN occurs in the presence of an electron acceptor such as phenazine methosulfate (Wissing, 1974).
In P. aeruginosa PAO1, the HCN synthase biosynthetic operon hcnABC was characterized (Pessi and Haas, 2000). hcnA was reported to encode a protein similar to formate dehydrogenase while hcnB and hcnC encode products with similarity to amino acid oxidases (Laville et al., 1998; Svercel et al., 2007). In a phylogenetic analysis of 30 fluorescent pseudomonads, no evidence was found for HGT of the hcn gene cluster, but rather that the locus appears to be exclusively inherited vertically (Frapolli et al., 2012). HCN has also been detected in Chromobacterium violaceum but the underlying genes have not been reported which might otherwise give new insights into HCN biosynthesis outside of the pseudomonads (Blom et al., 2011).
Bacteriocin
In this section, only the most well studied compound from this class is discussed. The structure of agrocin 84 described in this review is included (Figure 6).
Agrocin 84
Agrocin 84 is a 6-N-phosphoramidate of an adenine nucleotide analog (Roberts and Tate, 1977). This compound is produced by non-pathogenic strains of Agrobacterium radiobacter to biocontrol crown gall, a tumorous disease resulting from overproduction of auxin and cytokinin hormones stimulated by the Ti plasmid after it has transferred from A. radiobacter and integrated within host plant chromosomal DNA (Wang et al., 1994). Recently, it was shown that agrocin 84 employs a novel mechanism to inhibit leucyl-tRNA synthetases and hence inhibit translation (Chopra et al., 2013), though it was earlier suggested that agrocin 84 acts by inhibiting DNA synthesis (Das et al., 1978).
In Agrobacterium radiobacter K84, the biosynthesis and immunity to agrocin 84 is encoded by 17 coding sequences (the agn operon) located on a 44-kb conjugal plasmid, pAgK84, though the plasmid has 36 CDS in total (Kim et al., 2006). The two most interesting CDS are agnB2 and agnA which encode aminoacyl tRNA synthetase homologs. The agrocin 84 antibiotic is essentially a nucleotide attached to an amino acid-like moiety (methyl pentanamide), and its mode of action was proposed to involve competitive binding to the active site of leucyl-tRNA synthetases (Reader et al., 2005). agnB2 encodes a leucyl-tRNA synthetase homolog that confers self-immunity to agrocin 84 since it does not bind the antibiotic (Kim et al., 2006). Normally, a tRNA synthetase acts as a ligase that catalyzes the attachment of an amino acid to a tRNA which includes an anticodon; the catalysis results in a phosphoanhydride bond between the amino acid and ATP as the initial step in the aminoacylation of tRNA (Ibba and Söll, 2000). Surprisingly, agnA encodes a truncated homolog of an asparaginyl-tRNA synthetase which lacks the anticodon-binding domain, but maintains the catalytic domain. Thus, agnA appears to be a fascinating example of a gene that evolved from an ancient tRNA synthetase (for arginine), but now is a biosynthetic enzyme for an antibiotic that inhibits a paralogous enzyme (for leucine attachment) (Kim et al., 2006).
The agn operon may have an evolutionary history of horizontal gene transfer, as pAgK84 is inter and intra species transferable: Rhizobium that received the pAgK84 plasmid from Agrobacterium as trans-conjugates could synthesize agrocin 84 and received immunity as well (Farrand et al., 1985).
A final fascinating feature of the agn system is an apparent second form of ancient evolutionary pressure on the genes responsible for the biosynthesis of the antibiotic. Agrocin 84 is a chemical mimic of agrocinopines, a class of compounds that is a source of plant-derived nitrogen for the pathogens targeted by the antibiotic; the pathogens have their own Ti plasmids that encode for transporters that not only transport agrocinopines but also agrocin 84 (Ellis and Murphy, 1981; Hayman and Farrand, 1988; Kim and Farrand, 1997). Hence the agn biosynthetic genes evolved to create a chemical structure that not only mimics the tRNA synthetase substrate of the pathogen target, but also targets its nitrogen uptake machinery.
Enzymes
In this section, only the most well studied anti-fungal enzyme, chitinase, is discussed.
Chitinase
Chitinases are enzymes that break down chitin, one of the fungal cell wall components composed of repeated units of N-acetyl-D-glucos-2-amine, linked by β-1,4 glycosidic bonds (Bhattacharya et al., 2007). Fungi and hence chitin are enriched in soil and thus soil microbes are abundant sources of chitinases (also to target insects) (Hjort et al., 2010). Examples of chitinase-producing microbes include: fluorescent Pseudomonas strains isolated from the sugarcane rhizosphere that can target Colletotrichum falcatum, the causative agent of red rot disease in this crop (Viswanathan and Samiyappan, 2001); Actinoplanes missouriensis that antagonizes Plectosporium tabacinum, the causal agent of lupin root rot in Egypt (El-Tarabily, 2003); and Stenotrophomonas maltophilia that suppresses summer patch disease in Kentucky bluegrass (Kobayashi et al., 2002). Chitinases are produced by diverse bacterial genera including Pseudomonas, Streptomyces, Bacillus, and Burkholderia (Quecine et al., 2008). Chitinases are divided into two major categories, exochitinases and endochitinases. Of the four reported endochitinase family members (glycoside hydrolase families 18, 19, 23, and 48), primarily families 18 and 19 have been reported in bacteria, with only a single example of a family 23 chitinase (Prakash et al., 2010).
In Stenotrophomonas maltophilia 34S1, the chitinase family 18 gene has one CDS that encodes for a protein with seven domains: a catalytic domain, a chitin binding domain, three putative binding domains, a fibronectin type III domain and a polycystic kidney disease domain (Kobayashi et al., 2002). Bacterial chitinase family 18 has been shown to display different types of diversity. First, sequence analysis has shown that the catalytic domain and substrate binding domain, which are separated by a linker, have evolved independently. As the domain sequences do not match the taxonomies of their hosts, it has been suggested that domain swapping has been an important generator of diversity in this family, combined with HGT (Figure 7) (Karlsson and Stenlid, 2008). Unusual examples of chitinase genes are those that contain multiple family 18 catalytic domains within the same peptide that appear to function independently of one another (Howard et al., 2004). Additional examples of family 18 biodiversity include genes that contain non-consensus sequences at the catalytic site, as well as a bacterial subgroup that consists solely of a catalytic domain (Karlsson and Stenlid, 2008).
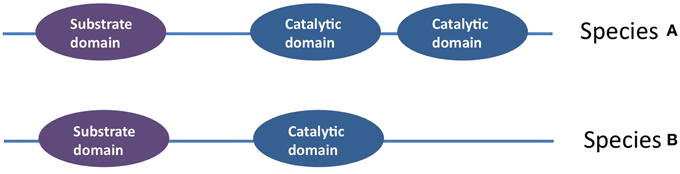
Figure 7. An example of intra- coding sequences diversification within an anti-microbial gene cluster: amongst the Family 18 chitinases is an example of a chitinase in which the catalytic domain has been duplicated (Howard et al., 2004).
Unlike family 18 chitinases that are widely distributed among the prokaryotes, family 19 chitinases are restricted to green non-sulfur and purple bacteria, as well as actinobacteria (Prakash et al., 2010). Based on sequence alignments of family 19 chitinases in prokaryotes and eukaryotes, strong evidence has emerged that this gene family in actinobacteria and purple bacteria was derived from flowering plants by HGT (Prakash et al., 2010). Furthermore, HGT from plants to purple bacteria may have occurred as two independent events in the distant past, followed more recently by HGT to actinobacteria (Prakash et al., 2010). The core architecture and catalytic sites of bacterial and plant family 19 chitinases are nearly identical. The sequence analysis further suggests that there was subsequent HGT from purple bacteria and actinobacteria to nematodes and arthropods, respectively (Prakash et al., 2010).
Discussion
The objective of this paper was to review the biodiversity of anti-microbial compounds, their mode(s) of action and underlying biosynthetic genes within plant associated microbes. This review covered diverse biosynthetic gene clusters that encode polyketides, non-ribosomal peptides, terpenoids, alkaloids, heterocyclic nitrogenous compounds, volatile compounds, bacteriocins, and lytic enzymes. The reviewed evidence suggests that these biosynthetic genes have diversified at different orders, each based on distinct evolutionary mechanisms:
Species Level Diversification
An emerging theme from the literature is that horizontal gene transfer (HGT) appears to have played a major role in the evolutionary diversification of plant-associated microbial species through inheritance of anti-microbial traits. There is evidence that HGT may have occurred from: bacteria to bacteria such as those that inhabit the rhizosphere (e.g., phenazine); from bacteria to fungi (e.g., helvolic acid); from bacteria to nematodes and arthropods (e.g., chitinase family 19); possibly from fungi to plants (e.g., Taxol); from plants to bacteria (e.g., phenazine and chitinase family 19); and even from higher eukaryotes to bacteria (e.g., IleS, pseudomonic acid resistance protein) (Figure 8). As noted in the literature, diverse factors might have facilitated these remarkable gene transfer events including: (1) the clustering of genes encoding the secondary metabolite; (2) homologous recombination between chromosomes and trans-conjugated plasmids (e.g., phenazine, zwittermicin A); (3) the presence of mobile elements (DNA transposons and retroelements) flanking the biosynthetic operons (e.g., zwittermicin A, phenazine and ergovaline); and (4) the presence of genes that encode self-immunity to the antibiotic within the biosynthetic cluster as otherwise receiving the cluster would have caused immediate suicide (e.g., mupirocin). It is worth noting that some gene clusters show no evidence of HGT (e.g., hydrogen cyanide).
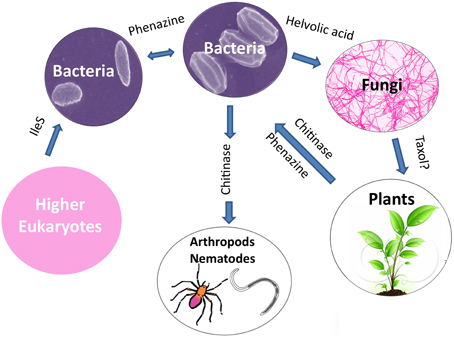
Figure 8. Potential examples of horizontal gene transfer of anti-microbial gene clusters leading to species level evolutionary diversification.
Genome Level Diversification
A second emerging theme from the literature is that a subset of associated plant- associated microbial genomes have diversified with respect to duplications of entire gene clusters responsible for the synthesis of antimicrobial compounds. For example, as noted above, in Neotyphodium uncinatum, there are two homologous gene clusters that encode loline, LOL-1 and LOL-2, the likely result of a segmental duplication event within this fungus. Another noted example is from Bacillus species in which three tandemly duplicated gene clusters, pks1, pks2, and pks3, encode the polyketides, bacillaene, macrolactin, and difficidin, respectively, the likely result of a homologous recombination event.
Intra Gene Cluster Diversification
A third interesting theme from the literature is that gene clusters encoding anti-microbial compounds have extensively diversified within, to permit biochemical diversification. The biosynthetic genes for these compounds are clustered in fungi or organized into operons in bacteria—in the latter, they are generally located on chromosomes but occasionally on plasmids (e.g., agrocin 84). Diversity within each cluster can include varying combinations of biosynthetic coding sequences (CDS), transporters for the respective compound, regulatory genes and CDS that confer self-immunity (e.g., mupirocin, agrocin 84). The biosynthetic operons vary in how many CDS synthesize the core skeleton (e.g., synthetases) as well as in how many encode decoration enzymes (e.g., hydroxylases, acyltransferases). However, the decoration enzymes may be encoded outside the gene cluster (e.g., phenazine operon). Furthermore, the biosynthetic CDS may be organized into genetic modules (e.g., NRPS) that vary in number. Each gene cluster is also associated with distinct DNA regulatory elements, for example to receive signals such as from quorum sensing. For example, comparative analysis of the trichothecene biosynthetic gene clusters (TRI) in Fusarium and Trichoderma showed multiple evolutionary diversification events within a single biosynthetic gene cluster family (e.g., head-to-tail vs. head-to-head rearrangements) (Figure 5). In another example, comparative analyses of the polymyxin operon showed mixing and matching of CDS, resulting in diversification of the compounds. Similarly, diversification of the phenazines likely arose through a diversity of biosynthetic decoration enzymes (e.g., hydroxylases). Another intriguing observation is from the helvolic acid biosynthetic gene cluster, in which transferase CDS were shown to have duplicated and diversified into paralogous gene families. As noted above, an interesting feature of this gene cluster is that it located in the sub-telomere chromosome region which is associated with high rates of evolutionary recombination.
Diversification Within Coding Sequences (CDS)
A final emerging theme from the literature is that diversification of anti-microbial traits in plant-associated microbes arose from allelic diversification. For example, intragenic swapping of domains was observed within the same genetic module (e.g., iturin A, mycosubtilins). As another example, whereas most chitinase genes possess a single catalytic domain, examples were noted where a single CDS encodes two catalytic domains (Figure 7). In general, the literature notes that domains within the same CDS can evolve independently (e.g., catalytic vs. substrate binding domains of chitinase); combined with the existence of linker peptides between domains as sites of homologous recombination, these features can result in novel alleles following domain swapping (e.g., family18 chitinases). Whereas, such allelic diversification plays a major role in the diversification of compound structures, caused for example by DNA mutations within the substrate binding domain, the literature demonstrates examples where biochemical diversity has arisen from relaxed substrate specificity of the biosynthetic enzymes (Figure 3). A representative example of the latter is the promiscuous fusaricidin NRPS in which the same recognition domain in different species can recognize and incorporate different amino acids, and furthermore it can recognize amino acids beyond the 21 standard amino acids translated by the ribosome, which results in significant structural diversity.
Dynamic Evolutionary Driven by Selection Pressures
It is interesting to speculate on the evolutionary selection pressures that have led to the diversification at the various biological levels noted above. At the most basic level, diversification was likely driven by a three-way co-evolution between the plant-associated microbe, its target pathogen and the host plant. This co-evolution may have occurred within a specific plant tissue niche or within soil associated with the rhizosphere (e.g., phenazine and PLt to combat soil-borne pathogens). However, there is also evidence for four-way interactions, to also include additional microbes (e.g., Taxol, jadomycin) and insects (e.g., ergovaline). These complex interactions can be bi-directional (e.g., loline). Within the producing organism, there is evidence for selection pressure to coordinate biosynthesis of the anti-microbial compound with the life cycle of the microbe (e.g., fusaricidins). There may also have been selection for genetic efficiency (e.g., potential sharing of transporter genes between polymyxin and fusaricidin). These selection pressures have led to fascinating individual stories, including the evolution of mimicry to facilitate antibiosis (e.g., agrocin).
Ecological and Evolutionary Lessons
When the examples of anti-microbial pathways were grouped by the phylogeny of their host microbes, several trends were observed (Figure 9, Tables 1, 2). Specifically: (1) some anti-microbial genes are apparently widely distributed among diverse taxonomic classes of bacteria (e.g., chitinases); (2) some metabolic pathways are widely distributed within one taxonomic class such as pyrrolnitrin that shows up in more than half of the presented species of Proteobacteria; (3) other anti-microbial pathways appear to be more restricted (e. g., fusaricidin, polymyxin, jadomycin). These results may correlate to the evolutionary age of these genetic pathways, or may represent a bias based on how well the pathway has been studied. More widespread genome sequencing and/or the use of orthologous gene probes may help to inform the evolutionary origins of these anti-microbial pathways.
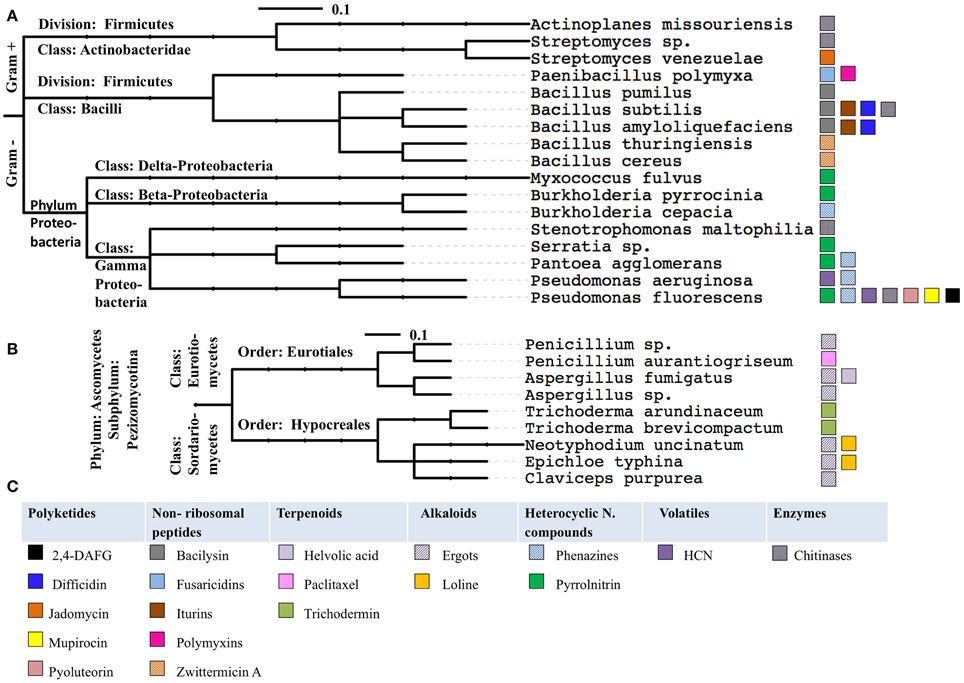
Figure 9. The anti-microbial compounds reviewed in this study grouped by the phylogeny of their microbial hosts, for bacteria (A) and fungi (B). The phylogenetic trees were generated using the interactive Tree of Life website (Letunic and Bork, 2011). The anti-microbial compounds produced by these species color coded, panel of color coded as indicated (C). The scale bar represents the number of nucleotide substitutions per site.
Bacterial Pathway Lessons
The selected examples of anti-microbial pathways from plant-associated bacteria found in the literature and presented in this review are distributed across Proteobacteria, Actinobacteria and Firmicutes (Figure 9). This may not be surprising as Proteobacteria and Actinobacteria are among the most widespread bacterial taxa associated with plants, perhaps because of their saprophytic capabilities (Bulgarelli et al., 2012).
Within these phyla, P. fluorescens (Proteobacteria) and B. subtilis (Firmicutes) were observed to produce a plethora of diverse antimicrobial compounds belonging to diverse chemical classes including polyketides, non-ribosomal peptides, heterocyclic nitrogenous compounds, volatiles and enzymes which reflect the diversity of the metabolic machineries of these species. As P. fluorescens and B. subtilis are both model systems, these results also support the above note of the bias within this literature.
Bacillus sp. and Pseudomonas sp. are ubiquitous microbes that can survive in diverse ecological niches (Compant et al., 2005). Both have elegant survival strategies that involve the production of antibiotics, surfactin, cyanide, biofilms, and induction of host resistance (Espinosa-Urgel, 2004; Dini-Andreote and van Elsas, 2013). These unique adaptations have led to their widespread study and use as biocontrol agents (Santoyo et al., 2012).
Genome analysis of P. fluorescens has provided insight into its ecological competency and evolutionary mechanisms. The versatile and rapid adaptability of P. fluorescens to diverse environmental clues may be attributed to over 200 characterized signal transduction proteins which enhance its sensing capability (Garbeva and de Boer, 2009; Humair et al., 2010). With respect to co-evolution, the P. fluorescens genome is exceptionally rich in repetitive extragenic palindromic (REP) elements, target sites for transposases and recombinases, with 1052 REP elements identified in P. fluorescens Pf-5 (compared to 21 in P. aeruginosa PAO1 and 365 in P. syringae DC 3000) (Tobes and Pareja, 2005, 2006). REPs likely affected genome evolution either by gene gain, loss or rearrangement (Silby et al., 2011). The latest version of the genome sequence and annotation of P. fluorescens was recently released (Martínez-García et al., 2015).
B. subtilis is naturally competent genetically, with a cascade of competence-specific DNA-uptake proteins that bind and transport DNA, in addition to a dynamic recombination mechanism which transforms chromosomal or plasmid DNA via different pathways (Chen and Dubnau, 2004; Kidane et al., 2009). Additionally, the B. subtilis genome encodes integrative and conjugative element binding (ICEBs1) proteins responsible for excision, integration, transfer of DNA (Lee et al., 2007) that likely have facilitated HGT. Comparative genomic analysis of B. subtilis strains revealed 298 accessory segments that potentially originated from mobile elements including plasmids, transposons and phages. This implies extensive HGT events that lead to diversification of the arsenal of anti-microbial pathways within Bacillus (Zeigler, 2011). The complete genome sequence and genome annotation of B. subtilis is available (Barbe et al., 2009; Belda et al., 2013).
Fungal Pathway Lessons
In contrast to bacteria, all the fungal examples presented in the review belong to a single classification—the Pezizomycotina (filamentous fungi), a subdivision of Ascomycota, the largest phylum of fungi (Blackwell, 2011) including representatives from Eurotiomycetes and Sordariomycetes (Figure 9). Pezizomycotina has an ancient origin in the Cambrian period, ca 530 Mya (Prieto and Wedin, 2013).
Pezizomycotina species are the most ubiquitous fungi with extremely diverse lifestyles, suggesting a corresponding diversity of ecological strategies (Spatafora et al., 2006; Beimforde et al., 2014) reflected in their production of a range of anti-microbial metabolites. There may be at least two reasons for this metabolic diversity, HGT and recombination. First, HGT from bacteria to fungi was previously reported in Ascomycota, of which 65% were observed in Pezizomycotina (Marcet-Houben and Gabaldón, 2010). Second, secondary metabolism gene clusters in Pezizomycotina show evidence of recent gene expansion (Arvas et al., 2007). Interestingly, most of these genes are located in the sub-telomere region (Rehmeyer et al., 2006) that is associated with a considerable high rate of recombination and correspondingly rapid evolution compared to other regions in the genome (Freitas-Junior et al., 2000), an example represented in this review by helvolic acid (Table 1).
Gaps and Future Perspectives
Despite the apparent progress in understanding the genetic mechanisms underlying the diversity of anti-microbial compounds produced by plant-associated microbes, significant gaps and opportunities remain. The major challenge is that a vast majority of plant associated microbes are unculturable, a phenomena that, to a far extent, limits our understanding of species diversification and evolution. It is worth noting that considerable progress toward cultivation of unculturable microbes has started to be achieved (Pham and Kim, 2012; Stewart, 2012). The modified cultivation methods attempt to simulate the natural environment, and include community culturing, and the use of high-throughput microbioreactors and laser microdissection (Pham and Kim, 2012). Another challenge is that the literature appears to be biased for model organisms, with insufficient data from other organisms in the phylogenetic tree for comparative genomics and evolutionary studies. For example, despite our efforts to originally focus this review only on anti-microbial pathways from endophytes, it became clear that the number of associated genes from endophytes has largely been unexplored, compared to free living rhizosphere model species. Indeed, there remains a lack of detailed genetic analysis underlying many anti-microbial compounds across microbes (endophytic and non-endophytic) and a lack of information to connect allelic diversity with compound diversity.
With respect to understanding the biosynthetic pathways of these metabolites, more information is needed as to the extent that diverse anti-microbial pathways coordinate and share biosynthetic enzymes. An important question in metabolic biosynthesis is understanding how chemical substrates are channeled along metabolic pathways from one enzyme to the next; from this review, it appears that some anti-microbial pathways solve this problem by using mega-synthase enzymes (e.g., zwittermicin A), but for other pathways, investigation of enzyme-enzyme interactions will be informative. An interesting future area of study will be to investigate the subcellular location of biosynthetic and storage proteins, especially of self-toxic compounds that may need to be sequestered. To that end, there have been advances in studying compartmentalization and secondary metabolite trafficking machinery (Roze et al., 2011; Lim and Keller, 2014; Kistler and Broz, 2015), which offer strategies to move forward.
A significant challenge in this discipline is the study of anti-microbial compounds in their native ecological context, as most reports are based only on in vitro studies. In particular, because the target pathogen affects the host plant, more information is needed as to how the plant and the anti-pathogenic microbe coordinate and regulate one another. For example, in the jadomycin pathway, evidence suggests that the plant sensing of the pathogen stimulates the anti-pathogen pathway in the associated beneficial microbe. The potential complexity of plant-microbe interactions and associated signaling networks are well studied in model systems such as Rhizobium (Janczarek et al., 2015). Though Rhizobium is a symbiotic microbe of legume plants, these studies suggest that a wealth of information remains to be explored for other plant-associated microbes, in particular endophytes (Kusari et al., 2012).
Also within the ecological context, basic biochemical questions are raised such as whether the anti-microbial pathway is regulated by the target pathogen, for example feedback inhibition once the pathogen has been eliminated. To help understand the genetic regulation of these anti-microbial pathways, analysis of gene expression with respect to the microbial life cycle would be a useful avenue of investigation, similar to the interesting findings from the fusaricidin pathway. An interesting study concerning aflatoxin, a polyketide mycotoxin, revealed strong evidence for the potential link between the fungal growth stage and polyketide biosynthesis (Zhou et al., 2000). Furthermore, intracellular tracking of aflatoxin biosynthetic enzymes in Aspergillus parasiticus showed significant accumulation in the vacuoles of specific cells but its absence in neighboring ones (Hong and Linz, 2008). This surprising result led Roze et al. (2011) to hypothesize the possibility of special and temporal gene expression of the associated biosynthetic pathway, at different developmental resolutions ranging from a single cell to fungal colony.
A related major challenge is that there are many natural products that exist in the literature that were initially isolated as part of screens for new compounds from total extracts, and hence the ecological functions of these compounds, as well as their underlying genes, remain unknown.
As anti-pathogenic metabolites may be self-toxic, the evolution of self-resistance is a particularly fascinating avenue of study, which this review demonstrates has been investigated for a limited number of pathways (e.g., mupirocin). Diverse self-resistance mechanisms have been reported in the microbial literature (Schäberle et al., 2011; Westman et al., 2013; Stegmann et al., 2015), suggesting that each plant-asssociated microbe with anti-microbial activity may employ unique self-protection strategies.
The recent advances in genome sequencing combined with gene editing tools will facilitate more in-depth analysis of orthologous biosynthetic genes in diverse species. Bioinformatic genome mining of biosynthetic gene clusters, combined with new advances in metabolomics, may also lead to the discovery of a diverse array of novel bio-active natural products. Moreover, merging these techniques with knowledge of microbial co-evolution and ecology (Vizcaino et al., 2014) along with advanced microscopy and imaging techniques will open a new era of discovery to harvest the diversity of natural products to combat evolving pathogens.
Author Contributions
WM wrote the manuscript, and WM and MR edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Travis Goron (University of Guelph) for helpful comments on the manuscript. WM was supported by a generous scholarship from the Government of Egypt. MR was supported by grants from the Grain Farmers of Ontario, OMAFRA, and the CIFSRF program by the International Development Research Centre and DFATD of the Government of Canada.
References
Anjaiah, V., Koedam, N., Nowak-Thompson, B., Loper, J. E., Höfte, M., Tambong, J. T., et al. (1998). Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives toward Fusarium spp. and Pythium spp. Mol. Plant Microbe Interact. 11, 847–854. doi: 10.1094/MPMI.1998.11.9.847
Arima, K., Imanaka, H., Kousaka, M., Fukuta, A., and Tamura, G. (1964). Pyrrolnitrin, a new antibiotic substance, produced by Pseudomonas. Agric. Biol. Chem. 28, 575–576. doi: 10.1271/bbb1961.28.575
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Arvas, M., Kivioja, T., Mitchell, A., Saloheimo, M., Ussery, D., Penttila, M., et al. (2007). Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina. BMC Genomics 8:325. doi: 10.1186/1471-2164-8-325
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Audenaert, K., Pattery, T., Cornelis, P., and Hofte, M. (2002). Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol. Plant Microbe Interact. 15, 1147–1156. doi: 10.1094/MPMI.2002.15.11.1147
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bangera, M. G., and Thomashow, L. S. (1999). Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 181, 3155–3163.
Barbe, V., Cruveiller, S., Kunst, F., Lenoble, P., Meurice, G., Sekowska, A., et al. (2009). From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155, 1758–1775. doi: 10.1099/mic.0.027839-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Barea, J. M., Pozo, M. J., Azcon, R., and Azcon-Aguilar, C. (2005). Microbial co-operation in the rhizosphere. J. Exp. Bot. 56, 1761–1778. doi: 10.1093/jxb/eri197
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Beatty, P. H., and Jensen, S. E. (2002). Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can. J. Microbiol. 48, 159–169. doi: 10.1139/w02-002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Beimforde, C., Feldberg, K., Nylinder, S., Rikkinen, J., Tuovila, H., Dörfelt, H., et al. (2014). Estimating the phanerozoic history of the Ascomycota lineages: combining fossil and molecular data. Mol. Phylogenet. Evol. 78, 386–398. doi: 10.1016/j.ympev.2014.04.024
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Belda, E., Sekowska, A., Le Fevre, F., Morgat, A., Mornico, D., Ouzounis, C., et al. (2013). An updated metabolic view of the Bacillus subtilis 168 genome. Microbiology 159, 757–770. doi: 10.1099/mic.0.064691-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bhattacharya, D., Nagpure, A., and Gupta, R. K. (2007). Bacterial chitinases: properties and potential. Crit. Rev. Biotechnol. 27, 21–28. doi: 10.1080/07388550601168223
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blackwell, M. (2011). The fungi: 1, 2, 3… 5.1 million species? Am. J. Bot. 98, 426–438. doi: 10.3732/ajb.1000298
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blankenship, J. D., Spiering, M. J., Wilkinson, H. H., Fannin, F. F., Bush, L. P., and Schardl, C. L. (2001). Production of loline alkaloids by the grass endophyte, Neotyphodium uncinatum, in defined media. Phytochemistry 58, 395–401. doi: 10.1016/S0031-9422(01)00272-2
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blom, D., Fabbri, C., Eberl, L., and Weisskopf, L. (2011). Volatile-mediated killing of Arabidopsis thaliana by bacteria is mainly due to hydrogen cyanide. Appl. Environ. Microbiol. 77, 1000–1008. doi: 10.1128/AEM.01968-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blom, J., Rueckert, C., Niu, B., Wang, Q., and Borriss, R. (2012). The complete genome of Bacillus amyloliquefaciens subsp. plantarum CAU B946 contains a gene cluster for nonribosomal synthesis of iturin A. J. Bacteriol. 194, 1845–1846. doi: 10.1128/JB.06762-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blumer, C., and Haas, D. (2000). Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173, 170–177. doi: 10.1007/s002039900127
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brandl, H., Lehmann, S., Faramarzi, M. A., and Martinelli, D. (2008). Biomobilization of silver, gold, and platinum from solid waste materials by HCN-forming microorganisms. Hydrometallurgy 94, 14–17. doi: 10.1016/j.hydromet.2008.05.016
Broderick, N. A., Goodman, R. M., Raffa, K. F., and Handelsman, J. (2000). Synergy between zwittermicin A and Bacillus thuringiensis subsp. kurstaki against gypsy moth (Lepidoptera: Lymantriidae). Environ. Entomol. 29, 101–107. doi: 10.1603/0046-225X-29.1.101
Bulgarelli, D., Rott, M., Schlaeppi, K., van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cane, D. E. (1997). A special thematic issue on polyketide and nonribosomal polypeptide biosynthesis. Chem. Rev. 97, 2463–2706. doi: 10.1021/cr970097g
Cardoza, R. E., Malmierca, M. G., Hermosa, M. R., Alexander, N. J., McCormick, S. P., Proctor, R. H., et al. (2011). Identification of loci and functional characterization of trichothecene biosynthesis genes in filamentous fungi of the genus Trichoderma. Appl. Environ. Microbiol. 77, 4867–4877. doi: 10.1128/AEM.00595-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carter, C. J., Cannon, M., and Smith, K. E. (1976). Inhibition of protein synthesis in Reticulocyte lysates by trichodermin. Biochem. J. 154, 171–178.
Castric, P. (1994). Influence of oxygen on the Pseudomonas aeruginosa hydrogen cyanide synthase. Curr. Microbiol. 29, 19–21. doi: 10.1007/BF01570186
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Castric, P. A. (1977). Glycine metabolism by Pseudomonas aeruginosa: hydrogen cyanide biosynthesis. J. Bacteriol. 130, 826–831.
Chen, I., and Dubnau, D. (2004). DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249. doi: 10.1038/nrmicro844
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, L., Chen, J., Zheng, X., Zhang, J., and Yu, X. (2007). Identification and antifungal activity of the metabolite of endophytic fungi isolated from Llex cornuta. Chin. J. Pestic. Sci. 9, 143–150.
Chen, X.-H., Scholz, R., Borriss, M., Junge, H., Mögel, G., Kunz, S., et al. (2009). Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 140, 38–44. doi: 10.1016/j.jbiotec.2008.10.015
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, X.-H., Vater, J., Piel, J., Franke, P., Scholz, R., Schneider, K., et al. (2006). Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188, 4024–4036. doi: 10.1128/JB.00052-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chernin, L., Brandis, A., Ismailov, Z., and Chet, I. (1996). Pyrrolnitrin production by an Enterobacter agglomerans strain with a broad spectrum of antagonistic activity towards fungal and bacterial phytopathogens. Curr. Microbiol. 32, 208–212. doi: 10.1007/s002849900037
Chin-A-Woeng, T. F., Bloemberg, G. V., van der Bij, A. J., van der Drift, K. M. G. M., Schripsema, J., Kroon, B., et al. (1998). Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. Radicis-lycopersici. Mol. Plant Microbe Interact. 11, 1069–1077. doi: 10.1094/MPMI.1998.11.11.1069
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Choi, S.-K., Park, S.-Y., Kim, R., Lee, C.-H., Kim, J. F., and Park, S. H. (2008). Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 365, 89–95. doi: 10.1016/j.bbrc.2007.10.147
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Choi, S.-K., Park, S.-Y., Kim, R., Kim, S.-B., Lee, C.-H., and Kim, J. F. (2009). Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J. Bacteriol. 191, 3350–3358. doi: 10.1128/JB.01728-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chopra, S., Palencia, A., Virus, C., Tripathy, A., Temple, B. R., Velazquez-Campoy, A., et al. (2013). Plant tumour biocontrol agent employs a tRNA-dependent mechanism to inhibit leucyl-tRNA synthetase. Nat. Commun. 4, 1417. doi: 10.1038/ncomms2421
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Compant, S., Duffy, B., Nowak, J., Clément, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Constantinescu, F. (2001). Extraction and identification of antifungal metabolites produced by some B. subtilis strains. Analele Institutului de Cercetari 31, 17–23.
Costa, R., van Aarle, I. M., Mendes, R., and Elsas, J. D. (2009). Genomics of pyrrolnitrin biosynthetic loci: evidence for conservation and whole-operon mobility within Gram-negative bacteria. Environ. Microbiol. 11, 159–175. doi: 10.1111/j.1462-2920.2008.01750.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Das, P. K., Basu, M., and Chatterjee, G. C. (1978). Studies on the mode of action of agrocin 84. J. Antibiot. 31, 490–492. doi: 10.7164/antibiotics.31.490
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Deane, C. D., and Mitchell, D. A. (2014). Lessons learned from the transformation of natural product discovery to a genome-driven endeavor. J. Ind. Microbiol. Biotechnol. 41, 315–331. doi: 10.1007/s10295-013-1361-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
de Groot, A. N., van Dongen, P. W., Vree, T. B., Hekster, Y. A., and van Roosmalen, J. (1998). Ergot Alkaloids. Drugs 56, 523–535. doi: 10.2165/00003495-199856040-00002
Delaney, S. M., Mavrodi, D. V., Bonsall, R. F., and Thomashow, L. S. (2001). phzO, a gene for biosynthesis of 2- hydroxylate phenazine compounds in Pseudomonas auerofacines. J. Bacteriol. 183, 5376–5384. doi: 10.1128/JB.183.1.318-327.2001
Delany, I., Sheehan, M. M., Fenton, A., Bardin, S., Aarons, S., and O'Gara, F. (2000). Regulation of production of the antifungal metabolite 2, 4-diacetylphloroglucinol in Pseudomonas fluorescens F113: genetic analysis of phlF as a transcriptional repressor. Microbiology 146, 537–546.
Dini-Andreote, F., and van Elsas, J. D. (2013). Back to the basics: the need for ecophysiological insights to enhance our understanding of microbial behaviour in the rhizosphere. Plant Soil 373, 1–15. doi: 10.1007/s11104-013-1687-z
Dotz, W. (1980). St. Anthony's fire. Am. J. Dermatopathol. 2, 249–254. doi: 10.1097/00000372-198000230-00013
Doull, J. L., Singh, A. K., Hoare, M., and Ayer, S. W. (1994). Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: effects of heat shock, ethanol treatment and phage infection. J. Ind. Microbiol. 13, 120–125. doi: 10.1007/BF01584109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dunlap, C. A., Bowman, M. J., and Schisler, D. A. (2013). Genomic analysis and secondary metabolite production in Bacillus amyloliquefaciens AS 43.3: a biocontrol antagonist of Fusarium head blight. Biol. Control 64, 166–175. doi: 10.1016/j.biocontrol.2012.11.002
Eilenberg, J. (2006). “Concepts and visions of biological control,” in An Ecological and Societal Approach to Biological Control, eds J. Eilenberg and H. M. T. Hokkanen (Dordrecht: Springer), 1–11.
El-Banna, N., and Winkelmann, G. (1998). Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 85, 69–78. doi: 10.1046/j.1365-2672.1998.00473.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellis, J., and Murphy, P. (1981). Four new opines from crown gall tumours - their detection and properties. Mol. Gen. Genet. 181, 36–43. doi: 10.1007/BF00339002
El-Sayed, A. K., Hothersall, J., Cooper, S. M., Stephens, E., Simpson, T. J., and Thomas, C. M. (2003). Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10, 419–430. doi: 10.1016/S1074-5521(03)00091-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
El-Sayed, A. K., Hothersall, J., and Thomas, C. M. (2001). Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology 147, 2127–2139.
El-Tarabily, K. A. (2003). An endophytic chitinase-producing isolate with potential for biological control of root rot of lupin caused by Plectosporium tabacinum. Aust. J. Bot. 51, 257–266. doi: 10.1071/BT02107
Eltringham, I. (1997). Mupirocin resistance and methicillin-resistant Staphylococcus aureus (MRSA). J. Hosp. Inf. 35, 1–8. doi: 10.1016/S0195-6701(97)90162-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Emmert, E. A., Klimowicz, A. K., Thomas, M. G., and Handelsman, J. (2004). Genetics of zwittermicin A production by Bacillus cereus. Appl. Environ. Microbiol. 70, 104–113. doi: 10.1128/AEM.70.1.104-113.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Espinosa-Urgel, M. (2004). Plant-associated Pseudomonas populations: molecular biology, DNA dynamics, and gene transfer. Plasmid 52, 139–150. doi: 10.1016/j.plasmid.2004.06.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Farrand, S. K., Slota, J. E., Shim, J. S., and Kerr, A. (1985). Tn5 insertions in the agrocin 84 plasmid: the conjugal nature of pAgK84 and the locations of determinants for transfer and agrocin 84 production. Plasmid 13, 106–117. doi: 10.1016/0147-619X(85)90063-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fleetwood, D. J., Scott, B., Lane, G. A., Tanaka, A., and Johnson, R. D. (2007). A complex ergovaline gene cluster in Epichloë endophytes of grasses. Appl. Environmen. Microbiol. 73, 2571–2579. doi: 10.1128/AEM.00257-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Flieger, M., Wurst, M., and Shelby, R. (1997). Ergot alkaloids-sources, structures and analytical methods. Folia Microbiol. 42, 3–30. doi: 10.1007/BF02898641
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Frapolli, M., Pothier, J. F., Défago, G., and Moënne, Y. (2012). Evolutionary history of synthesis pathway genes for phloroglucinol and cyanide antimicrobials in plant-associated fluorescent pseudomonads. Mol. Phylogen. Evol. 63, 877–890. doi: 10.1016/j.ympev.2012.02.030
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Freitas-Junior, L. H., Bottius, E., Pirrit, L. A., Deitsch, K. W., Scheidig, C., Guinet, F., et al. (2000). Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407, 1018–1022. doi: 10.1038/35039531
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fuller, A., Mellows, G., Woolford, M., Banks, G., Barrow, K., and Chain, E. (1971). Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature 234, 416–417. doi: 10.1038/234416a0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gales, A., Jones, R., and Sader, H. (2006). Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin. Microbiol. Infect. 12, 315–321. doi: 10.1111/j.1469-0691.2005.01351.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Garbeva, P., and de Boer, W. (2009). Inter-specific interactions between carbon-limited soil bacteria affect behavior and gene expression. Microb. Ecol. 58, 36–46. doi: 10.1007/s00248-009-9502-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ge, Y.-H., Pei, D.-L., Zhao, Y.-H., Li, W.-W., Wang, S.-F., and Xu, Y.-Q. (2007). Correlation between antifungal agent phenazine-1-carboxylic acid and pyoluteorin biosynthesis in Pseudomonas sp. m18. Curr. Microbiol. 54, 277–281. doi: 10.1007/s00284-006-0317-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Giddens, S. R., Feng, Y., and Mahanty, H. K. (2002). Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol. Microbiol. 45, 769–783. doi: 10.1046/j.1365-2958.2002.03048.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gröger, D., and Floss, H. G. (1998). “Biochemistry of ergot alkaloids - achievements and challenges,” in The Alkaloids: Chemistry and Biology, ed G. A. Cordell (New York, NY: Academic Press), 171–218.
Gueldner, R. C., Reilly, C. C., Pusey, P. L., Costello, C. E., Arrendale, R. F., Cox, R. H., et al. (1988). Isolation and identification of iturins as antifungal peptides in biological control of peach brown rot with Bacillus subtilis. J. Agric. Food Chem. 36, 366–370. doi: 10.1021/jf00080a031
Gurney, R., and Thomas, C. M. (2011). Mupirocin: biosynthesis, special features and applications of an antibiotic from a Gram-negative bacterium. Appl. Microbiol. Biotechnol. 90, 11–21. doi: 10.1007/s00253-011-3128-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gurusiddaiah, S., Weller, D., Sarkar, A., and Cook, R. (1986). Characterization of an antibiotic produced by a strain of Pseudomonas fluorescens inhibitory to Gaeumannomyces graminis var. tritici and Pythium spp. Antimicrob. Agent Chemother. 29, 488–495. doi: 10.1128/AAC.29.3.488
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Haarmann, T., Lorenz, N., and Tudzynski, P. (2008). Use of a nonhomologous end joining deficient strain (Deltaku70) of the ergot fungus Claviceps purpurea for identification of a nonribosomal peptide synthetase gene involved in ergotamine biosynthesis. Fungal Genet. Biol. 45, 35–44. doi: 10.1016/j.fgb.2007.04.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Haarmann, T., Machado, C., Lübbe, Y., Correia, T., Schardl, C. L., Panaccione, D. G., et al. (2005). The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry 66, 1312–1320. doi: 10.1016/j.phytochem.2005.04.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hamdache, A., Azarken, R., Lamarti, A., Aleu, J., and Collado, I. (2013). Comparative genome analysis of Bacillus spp. and its relationship with bioactive nonribosomal peptide production. Phytochem. Rev. 12, 685–716. doi: 10.1007/s11101-013-9278-4
Hammer, P. E., Burd, W., Hill, D. S., Ligon, J. M., and van Pee, K.-H. (1999). Conservation of the pyrrolnitrin biosynthetic gene cluster among six pyrrolnitrin-producing strains. FEMS Microbiol. Lett. 180, 39–44. doi: 10.1111/j.1574-6968.1999.tb08775.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hammer, P. E., Hill, D. S., Lam, S. T., van Pée, K. H., and Ligon, J. M. (1997). Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63, 2147–2154.
Hammer, P., Evensen, K., and Janisiewicz, W. (1993). Postharvest control of Botrytis cinerea on cut rose flowers with pyrrolnitrin. Plant Dis. 77, 283–286. doi: 10.1094/PD-77-0283
Han, J. W., Kim, E. Y., Lee, J. M., Kim, Y. S., Bang, E., and Kim, B. S. (2012). Site-directed modification of the adenylation domain of the fusaricidin nonribosomal peptide synthetase for enhanced production of fusaricidin analogs. Biotechnol. Lett. 34, 1327–1334. doi: 10.1007/s10529-012-0913-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Han, L., Yang, K., Ramalingam, E., Mosher, R. H., and Vining, L. C. (1994). Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuelae ISP5230. Microbiology 140, 3379–3389. doi: 10.1099/13500872-140-12-3379
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hassan, H. M., and Fridovich, I. (1980). Mechanism of the antibiotic action pyocyanine. J. Bacteriol. 141, 156–163.
Hassett, D. J., Schweizer, H. P., and Ohman, D. E. (1995). Pseudomonas aeruginosa sodA and sodB mutants defective in manganese-and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J. Bacteriol. 177, 6330–6337.
Hayman, G., and Farrand, S. (1988). Characterization and mapping of the agrocinopine-agrocin 84 locus on the nopaline Ti plasmid pTiC58. J. Bacteriol. 170, 1759–1767.
Heinig, U., Scholz, S., and Jennewein, S. (2013). Getting to the bottom of taxol biosynthesis by fungi. Fungal Divers. 60, 161–170. doi: 10.1007/s13225-013-0228-7
Heinstein, P. F., Lee, S. I., and Floss, H. G. (1971). Isolation of dimethylallylpyrophosphate: tryptophan dimethylallyl transferase from the ergot fungus (Claviceps spec.). Biochem. Biophys. Res. Commun. 44, 1244–1251. doi: 10.1016/S0006-291X(71)80219-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hezari, M., Lewis, N. G., and Croteau, R. (1995). Purification and characterization of taxa-4(5),11(12)-diene synthase from pacific yew (Taxus brevifolia) that catalyzes the first committed step of taxol biosynthesis. Arch. Biochem. Biophys. 322, 437–444. doi: 10.1006/abbi.1995.1486
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hilton, M. D., Alaeddinoglu, N. G., and Demain, A. L. (1988). Synthesis of bacilysin by Bacillus subtilis branches from prephenate of the aromatic amino acid pathway. J. Bacteriol. 170, 482–484.
Hjort, K., Bergström, M., Adesina, M. F., Jansson, J. K., Smalla, K., and Sjöling, S. (2010). Chitinase genes revealed and compared in bacterial isolates, DNA extracts and a metagenomic library from a phytopathogen suppressive soil. FEMS Microbiol. Ecol. 71, 197–207. doi: 10.1111/j.1574-6941.2009.00801.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hohn, T. M., and Beremand, P. D. (1989). Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79, 131–138. doi: 10.1016/0378-1119(89)90098-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hong, S. Y., and Linz, J. E. (2008). Functional expression and subcellular localization of the aflatoxin pathway enzyme Ver-1 fused to enhanced green fluorescent protein. Appl. Environ. Microbiol. 74, 6385–6396. doi: 10.1128/AEM.01185-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Howard, M. B., Ekborg, N. A., Taylor, L. E., Weiner, R. M., and Hutcheson, S. W. (2004). Chitinase B of “microbulbifer degradans” 2-40 contains two catalytic domains with different chitinolytic activities. J. Bacteriol. 186, 1297–1303. doi: 10.1128/JB.186.5.1297-1303.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Howell, C. R., and Stipanovic, R. D. (1980). Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70, 712–715. doi: 10.1094/Phyto-70-712
Hughes, J., and Mellows, G. (1980). Interaction of pseudomonic acid A with Escherichia coli B isoleucyl-tRNA synthetase. Biochem. J. 191, 209–219.
Humair, B., Wackwitz, B., and Haas, D. (2010). GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 76, 1497–1506. doi: 10.1128/AEM.02014-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ibba, M., and Söll, D. (2000). Aminoacyl-tRNA synthesis. Ann. Rev. Biochem. 69, 617–650. doi: 10.1146/annurev.biochem.69.1.617
Inaoka, T., Takahashi, K., Ohnishi-Kameyama, M., Yoshida, M., and Ochi, K. (2003). Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278, 2169–2176. doi: 10.1074/jbc.M208722200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Inaoka, T., Wang, G., and Ochi, K. (2009). ScoC regulates bacilysin production at the transcription level in Bacillus subtilis. J. Bacteriol. 191, 7367–7371. doi: 10.1128/JB.01081-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jakeman, D. L., Bandi, S., Graham, C. L., Reid, T. R., Wentzell, J. R., and Douglas, S. E. (2009). Antimicrobial activities of jadomycin B and structurally related analogues. Antimicrob. Agent Chemother. 53, 1245–1247. doi: 10.1128/AAC.00801-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Janczarek, M., Rachwał, K., Marzec, A., Grzą, J., and Palusińska-Szysz, M. (2015). Signal molecules and cell-surface components involved in early stages of the legume–rhizobium interactions. Appl. Soil Ecol. 85, 94–113. doi: 10.1016/j.apsoil.2014.08.010
Johnston-Monje, D., and Raizada, M. N. (2011). Conservation and diversity of seed associated endophytes in across boundaries of evolution, ethnography and ecology. PLoS ONE. 6:e20396. doi: 10.1371/journal.pone.0020396
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kajimura, Y., and Kaneda, M. (1996). Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8. Taxonomy, fermentation, isolation, structure elucidation and biological activity. J. Antibiot. 49, 129–135. doi: 10.7164/antibiotics.49.129
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kajimura, Y., and Kaneda, M. (1997). Fusaricidins B, C and D, new depsipeptide antibiotics produced by Bacillus polymyxa KT-8: isolation, structure elucidation and biological activity. J. Antibiot. 50, 220–228. doi: 10.7164/antibiotics.50.220
Karlsson, M., and Stenlid, J. (2008). Evolution of family 18 glycoside hydrolases: diversity, domain structures and phylogenetic relationships. J. Mol. Microbiol. Biotechnol. 16, 208–223. doi: 10.1159/000151220
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keller, L., and Surette, M. G. (2006). Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258. doi: 10.1038/nrmicro1383
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism-from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kenig, M., and Abraham, E. (1976). Antimicrobial activities and antagonists of bacilysin and anticapsin. J. Gen. Microbiol. 94, 37–45. doi: 10.1099/00221287-94-1-37
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kenig, M., vandamme, E. T., and Abraham, E. (1976). The mode of action of bacilysin and anticapsin and biochemical properties of bacilysin-resistant mutants. J. Gen. Microbiol. 94, 46–54. doi: 10.1099/00221287-94-1-46
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kevany, B. M., Rasko, D. A., and Thomas, M. G. (2009). Characterization of the complete zwittermicin A biosynthesis gene cluster from Bacillus cereus. Appl. Environ. Microbiol. 75, 1144–1155. doi: 10.1128/AEM.02518-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kidane, D., Carrasco, B., Manfredi, C., Rothmaier, K., Ayora, S., Tadesse, S., et al. (2009). Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells. PLoS Genet. 5:e1000630. doi: 10.1371/journal.pgen.1000630
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, B.-Y., Lee, S.-Y., Ahn, J.-H., Song, J., Kim, W.-G., and Weon, H.-Y. (2015). Complete genome sequence of Bacillus amyloliquefaciens subsp. plantarum CC178, a phyllosphere bacterium antagonistic to plant pathogenic fungi. Genome Announc. 3, e01368–e01314. doi: 10.1128/genomeA.01368-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, H., and Farrand, S. K. (1997). Characterization of the acc operon from the nopaline-type Ti plasmid pTiC58, which encodes utilization of agrocinopines A and B and susceptibility to agrocin 84. J. Bacteriol. 179, 7559–7572.
Kim, J.-G., Park, B. K., Kim, S.-U., Choi, D., Nahm, B. H., Moon, J. S., et al. (2006). Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the trojan horse antibiotic that controls crown gall. Proc. Natl. Acad. Sci. U.S.A. 103, 8846–8851. doi: 10.1073/pnas.0602965103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kirner, S., Hammer, P. E., Hill, D. S., Altmann, A., Fischer, I., Weislo, L. J., et al. (1998). Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180, 1939–1943.
Kistler, H., and Broz, K. (2015). Cellular compartmentalization of secondary metabolism. Front. Microbiol. 6:68. doi: 10.3389/fmicb.2015.00068
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kobayashi, D. Y., Reedy, R. M., Bick, J., and Oudemans, P. V. (2002). Characterization of a chitinase gene from Stenotrophomonas maltophilia strain 34S1 and its involvement in biological control. Appl. Environ. Microbiol. 68, 1047–1054. doi: 10.1128/AEM.68.3.1047-1054.2002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Köroğlu, T. E., Öğülür, İ., Mutlu, S., Yazgan-Karataş, A., and Özcengiz, G. (2011). Global regulatory systems operating in bacilysin biosynthesis in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 20, 144–155. doi: 10.1159/000328639
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kraus, J., and Loper, J. E. (1995). Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 61, 849–854.
Kusari, S., Hertweck, C., and Spiteller, M. (2012). Chemical ecology of endophytic fungi: origins of secondary metabolites. Chem. Biol. 19, 792–798. doi: 10.1016/j.chembiol.2012.06.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Laville, J., Blumer, C., Von Schroetter, C., Gaia, V., Défago, G., Keel, C., et al. (1998). Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 180, 3187–3196.
Leclère, V., Béchet, M., Adam, A., Guez, J., Wathelet, B., Ongena, M., et al. (2005). Mycosubtilin overproduction by Bacillus subtilis BBG100 enhances the organism's antagonistic and biocontrol activities. Appl. Environ. Microbiol. 71, 4577–4584. doi: 10.1128/AEM.71.8.4577-4584.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, C. A., Auchtung, J. M., Monson, R. E., and Grossman, A. D. (2007). Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 66, 1356–1369. doi: 10.1111/j.1365-2958.2007.06000.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leoffler, W., Tschen, J., Venittanakom, N., Kugler, M., Knorpp, E., Hsieh, T. F., et al. (1986). Antifungal effects of bacilysin and fengycin from Bacillus subtilis F-29-3: a comparison with activaties of other Bacillus antibiotics. J. Phytopathol. 115, 204–213. doi: 10.1111/j.1439-0434.1986.tb00878.x
Letunic, I., and Bork, P. (2011). Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucl. Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ligon, J. M., Hill, D. S., Hammer, P. E., Torkewitz, N. R., Hofmann, D., Kempf, H. J., et al. (2000). Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag. Sci. 56, 688–695. doi: 10.1002/1526-4998(200008)56:8
Li, J., and Jensen, S. E. (2008). Nonribosomal biosynthesis of fusaricidins by Paenibacillus polymyxa pkb1 involves direct activation of a d-amino acid. Chem. Biol. 15, 118–127. doi: 10.1016/j.chembiol.2007.12.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. doi: 10.1016/S1473-3099(06)70580-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, S., Zhang, R., Wang, Y., Zhang, N., Shao, J., Qiu, M., et al. (2013). Promoter analysis and transcription regulation of fus gene cluster responsible for fusaricidin synthesis of Paenibacillus polymyxa SQR-21. Appl. Microbiol. Biotechnol. 97, 9479–9489. doi: 10.1007/s00253-013-5157-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lim, F. Y., and Keller, N. P. (2014). Spatial and temporal control of fungal natural product synthesis. Nat. Prod. Rep. 31, 1277–1286. doi: 10.1039/C4NP00083H
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lodeiro, S., Xiong, Q., Wilson, W. K., Ivanova, Y., Smith, M. L., May, G. S., et al. (2009). Protostadienol biosynthesis and metabolism in the pathogenic fungus Aspergillus fumigatus. Org. Lett. 11, 1241–1244. doi: 10.1021/ol802696a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lu, J., Huang, X., Li, K., Li, S., Zhang, M., Wang, Y., et al. (2009). LysR family transcriptional regulator PqsR as repressor of pyoluteorin biosynthesis and activator of phenazine-1-carboxylic acid biosynthesis in Pseudomonas sp. M18. J. Biotechnol. 143, 1–9. doi: 10.1016/j.jbiotec.2009.06.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Luo, Y., Ruan, L.-F., Zhao, C.-M., Wang, C.-X., Peng, D.-H., and Sun, M. (2011). Validation of the intact zwittermicin A biosynthetic gene cluster and discovery of a complementary resistance mechanism in Bacillus thuringiensis. Antimicrob. Agent Chemother. 55, 4161–4169. doi: 10.1128/AAC.00111-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marcet-Houben, M., and Gabaldón, T. (2010). Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26, 5–8. doi: 10.1016/j.tig.2009.11.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martínez-García, P. M., Ruano-Rosa, D., Schilirò, E., Prieto, P., Ramos, C., Rodríguez-Palenzuela, P., et al. (2015). Complete genome sequence of Pseudomonas fluorescens strain PICF7, an indigenous root endophyte from olive (Olea europaea L.) and effective biocontrol agent against Verticillium dahliae. Stand. Genomic Sci. 10:10. doi: 10.1186/1944-3277-10-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mavrodi, D. V., Blankenfeldt, W., and Thomashow, L. S. (2006). Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44, 417–445. doi: 10.1146/annurev.phyto.44.013106.145710
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mavrodi, D. V., Parejko, J. A., Mavrodi, O. V., Kwak, Y., Weller, D. M., Blankenfeldt, W., et al. (2013). Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol. 15, 675–686. doi: 10.1111/j.1462-2920.2012.02846.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mavrodi, D. V., Peever, T. L., Mavrodi, O. V., Parejko, J. A., Raaijmakers, J. M., Lemanceau, P., et al. (2010). Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 76, 866–879. doi: 10.1128/AEM.02009-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mavrodi, O. V., McSpadden, G. B. B., Mavrodi, D. V., Bonsall, R. F., Weller, D. M., and Thomashow, L. S. (2001). Genetic diversity of phlD from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology 91, 35–43. doi: 10.1094/PHYTO.2001.91.1.35
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mazzola, M., Cook, R. J., Thomashow, L. S., Weller, D., and Pierson, L. (1992). Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58, 2616–2624.
McDonald, M., Mavrodi, D. V., Thomashow, L. S., and Floss, H. G. (2001). Phenazine biosynthesis in Pseudomonas fluorescens: branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. J. Am. Chem. Soc. 123, 9459–9460. doi: 10.1021/ja011243+
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McSpadden Gardener, B. B., Schroeder, K. L., Kalloger, S. E., Raaijmakers, J. M., Thomashow, L. S., and Weller, D. M. (2000). Genotypic and phenotypic diversity of phlD containing Pseudomonas strains isolated from the rhizosphere of wheat. Appl. Environ. Microbiol. 66, 1939–1946. doi: 10.1128/AEM.66.5.1939-1946.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Moule, Y., Jemmali, M., and Rousseau, N. (1976). Mechanism of the inhibition of transcription by pr toxin, a mycotoxin from Penicillium roqueforti. Chem. Biol. Interact. 14, 207–216. doi: 10.1016/0009-2797(76)90101-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mousa, W. K., and Raizada, M. N. (2013). The diversity of anti-microbial secondary metabolites produced by fungal endophytes: an interdisciplinary perspective. Front. Microbiol. 4:65. doi: 10.3389/fmicb.2013.00065
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nodwell, J. R. (2014). Are you talking to me? a possible role for γ butyrolactones in interspecies signalling. Mol. Microbiol. 94, 483–485. doi: 10.1111/mmi.12787
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nowak-Thompson, B., Chaney, N., Wing, J. S., Gould, S. J., and Loper, J. E. (1999). Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 181, 2166–2174.
Parker, J. B., and Walsh, C. T. (2012). Olefin isomerization regiochemistries during tandem action of bacA and bacB on prephenate in bacilysin biosynthesis. Biochemistry 51, 3241–3251. doi: 10.1021/bi300254u
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pessi, G., and Haas, D. (2000). Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182, 6940–6949. doi: 10.1128/JB.182.24.6940-6949.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pham, V. H., and Kim, J. (2012). Cultivation of unculturable soil bacteria. Trends Biotechnol. 30, 475–484. doi: 10.1016/j.tibtech.2012.05.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Phister, T. G., O'Sullivan, D. J., and McKay, L. L. (2004). Identification of bacilysin, chlorotetaine, and iturin a produced by Bacillus sp. strain cs93 isolated from pozol, a Mexican fermented maize dough. Appl. Environ. Microbiol. 70, 631–634. doi: 10.1128/AEM.70.1.631-634.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Picard, C., Di Cello, F., Ventura, M., Fani, R., and Guckert, A. (2000). Frequency and biodiversity of 2, 4-diacetylphloroglucinol-producing bacteria isolated from the maize rhizosphere at different stages of plant growth. Appl. Environ. Microbiol. 66, 948–955. doi: 10.1128/AEM.66.3.948-955.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pierson, I. L. S., and Pierson, E. A. (2010). Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 86, 1659–1670. doi: 10.1007/s00253-010-2509-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pierson, I. L., Wood, D., Pierson, E., and Chancey, S. (1998). N-acyl-homoserine lactone-mediated gene regulation in biological control by fluorescent pseudomonads: current knowledge and future work. Eur. J. Plant Pathol. 104, 1–9. doi: 10.1023/A:1008674417007
Prakash, N. U., Jayanthi, M., Sabarinathan, R., Kangueane, P., Mathew, L., and Sekar, K. (2010). Evolution, homology conservation, and identification of unique sequence signatures in GH19 family chitinases. J. Mol. Evol. 70, 466–478. doi: 10.1007/s00239-010-9345-z
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Prieto, M., and Wedin, M. (2013). Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS ONE 8:e65576. doi: 10.1371/journal.pone.0065576
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Proctor, R. H., and Hohn, T. M. (1993). Aristolochene synthase. isolation, characterization, and bacterial expression of a sesquiterpenoid biosynthetic gene (Ari1) from Penicillium roqueforti. J. Biol. Chem. 268, 4543–4548.
Proctor, R. H., McCormick, S. P., Alexander, N. J., and Desjardins, A. E. (2009). Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 74, 1128–1142. doi: 10.1111/j.1365-2958.2009.06927.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Quecine, M. C., Araujo, W. L., Marcon, J., Gai, C. S., Azevedo, J. L., and Pizzirani-Kleiner, A. A. (2008). Chitinolytic activity of endophytic Streptomyces and potential for biocontrol. Lett. Appl. Microbiol. 47, 486–491. doi: 10.1111/j.1472-765X.2008.02428.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Raffel, S. J., Stabb, E. V., Milner, J. L., and Handelsman, J. (1996). Genotypic and phenotypic analysis of zwittermicin A-producing strains of Bacillus cereus. Microbiology 142, 3425–3436. doi: 10.1099/13500872-142-12-3425
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rajavel, M., Mitra, A., and Gopal, B. (2009). Role of Bacillus subtilis BacB in the synthesis of bacilysin. J. Biol. Chem. 284, 31882–31892. doi: 10.1074/jbc.M109.014522
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reader, J. S., Ordoukhanian, P. T., Kim, J.-G., de Crécy-Lagard, V., Hwang, I., Farrand, S., et al. (2005). Major biocontrol of plant tumors targets tRNA synthetase. Science 309, 1533–1533. doi: 10.1126/science.1116841
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rehmeyer, C., Li, W., Kusaba, M., Kim, Y.-S., Brown, D., Staben, C., et al. (2006). Organization of chromosome ends in the rice blast fungus, Magnaporthe oryzae. Nucl. Acids Res. 34, 4685–4701. doi: 10.1093/nar/gkl588
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Roberts, W., and Tate, M. (1977). Agrocin 84 is a 6-N-phosphoramidate of an adenine nucleotide analogue. Nature 265, 379–381. doi: 10.1038/265379a0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Roze, L. V., Chanda, A., and Linz, J. E. (2011). Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet. Biol. 48, 35–48. doi: 10.1016/j.fgb.2010.05.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santoyo, G., Orozco-Mosqueda, M. D. C., and Govindappa, M. (2012). Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Bio. Sci. Technol. 22, 855–872. doi: 10.1080/09583157.2012.694413
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schäberle, T. F., Vollmer, W., Frasch, H.-J., Hüttel, S., Kulik, A., Röttgen, M., et al. (2011). Self-resistance and cell wall composition in the glycopeptide producer Amycolatopsis balhimycina. Antimicrob. Agent. Chemother. 55, 4283–4289. doi: 10.1128/AAC.01372-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schardl, C. L. (2010). The epichloae, symbionts of the grass subfamily poöideae1. Ann. Mo. Bot. Gard. 97, 646–665. doi: 10.3417/2009144
Scheffler, R., Colmer, S., Tynan, H., Demain, A., and Gullo, V. (2013). Antimicrobials, drug discovery, and genome mining. Appl. Microbiol. Biotechnol. 97, 969–978. doi: 10.1007/s00253-012-4609-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schneider, K., Chen, X.-H., Vater, J., Franke, P., Nicholson, G., Borriss, R., et al. (2007). Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 70, 1417–1423. doi: 10.1021/np070070k
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwarzer, D., Finking, R., and Marahiel, M. A. (2003). Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20, 275–287. doi: 10.1039/b111145k
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shaheen, M., Li, J., Ross, A. C., Vederas, J. C., and Jensen, S. E. (2011). Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem. Biol. 18, 1640–1648. doi: 10.1016/j.chembiol.2011.09.017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shanahan, P., Borro, A., O'Gara, F., and Glennon, J. D. (1992). Isolation, trace enrichment and liquid chromatographic analysis of diacetylphloroglucinol in culture and soil samples using UV and amperometric detection. J. Chromatogr. 606, 171–177. doi: 10.1016/0021-9673(92)87022-Z
Silby, M. W., Winstanley, C., Godfrey, S. A., Levy, S. B., and Jackson, R. W. (2011). Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680. doi: 10.1111/j.1574-6976.2011.00269.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Silo-Suh, L. A., Stabb, E. V., Raffel, S. J., and Handelsman, J. (1998). Target range of zwittermicin A, an aminopolyol antibiotic from Bacillus cereus. Curr. Microbiol. 37, 6–11. doi: 10.1007/s002849900328
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Silva, G. H., de Oliveira, C. M., Teles, H. L., Pauletti, P. M., Castro-Gamboa, I., Silva, D. H. S., et al. (2010). Sesquiterpenes from Xylaria sp., an endophytic fungus associated with Piper aduncum (Piperaceae). Phytochem. Lett. 3, 164–167. doi: 10.1016/j.phytol.2010.07.001
Soliman, S., and Raizada, M. N. (2013). Interactions between co-habitating fungi elicit synthesis of Taxol from an endophytic fungus in host Taxus plants. Front. Microbiol. 4:3. doi: 10.3389/fmicb.2013.00003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Soliman, S., Trobacher, C., Tsao, R., Greenwood, J., and Raizada, M. N. (2013). A fungal endophyte induces transcription of genes encoding a redundant fungicide pathway in its host plant. BMC Plant Biol. 13:93. doi: 10.1186/1471-2229-13-93
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Souza, J. T., and Raaijmakers, J. M. (2003). Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin−producing Pseudomonas and Burkholderia spp. FEMS Microbiol. Ecol. 43, 21–34. doi: 10.1111/j.1574-6941.2003.tb01042.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spatafora, J. W., Sung, G.-H., Johnson, D., Hesse, C., O'Rourke, B., Serdani, M., et al. (2006). A five-gene phylogeny of Pezizomycotina. Mycologia 98, 1018–1028. doi: 10.3852/mycologia.98.6.1018
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spiering, M. J., Moon, C. D., Wilkinson, H. H., and Schardl, C. L. (2005). Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 169, 1403–1414. doi: 10.1534/genetics.104.035972
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stegmann, E., Frasch, H.-J., Kilian, R., and Pozzi, R. (2015). Self-resistance mechanisms of actinomycetes producing lipid II-targeting antibiotics. Int. J. Med. Microbiol. 305, 190–195. doi: 10.1016/j.ijmm.2014.12.015
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stein, T. (2005). Bacillus subtilis antibiotics: structures, synthesis and specific functions. Mol. Microbiol. 56, 845–857. doi: 10.1111/j.1365-2958.2005.04587.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Steinborn, G., Hajirezaei, M.-R., and Hofemeister, J. (2005). bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains. Arch. Microbiol. 183, 71–79. doi: 10.1007/s00203-004-0743-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stewart, E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi: 10.1128/JB.00345-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Storm, D. R., Rosenthal, K. S., and Swanson, P. E. (1977). Polymyxin and related peptide antibiotics. Ann. Rev. Biochem. 46, 723–763. doi: 10.1146/annurev.bi.46.070177.003451
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sutherland, R., Boon, R., Griffin, K., Masters, P., Slocombe, B., and White, A. (1985). Antibacterial activity of mupirocin (pseudomonic acid), a new antibiotic for topical use. Antimicrob. Agents Chemother. 27, 495–498. doi: 10.1128/AAC.27.4.495
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Svercel, M., Duffy, B., and Defago, G. (2007). PCR amplification of hydrogen cyanide biosynthetic locus hcnaB in Pseudomonas spp. J. Microbiol. Methods 70, 209–213. doi: 10.1016/j.mimet.2007.03.018
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Takeno, A., Okamoto, A., Tori, K., Oshima, K., Hirakawa, H., Toh, H., et al. (2012). Complete genome sequence of Bacillus cereus NC7401, which produces high levels of the emetic toxin cereulide. J. Bacteriol. 194, 4767–4768. doi: 10.1128/JB.01015-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tazawa, J., Watanabe, K., Yoshida, H., Sato, M., and Homma, Y. (2000). Simple method of detection of the strains of fluorescent Pseudomonas spp. producing antibiotics, pyrrolnitrin and phloroglucinol. Soil Microorg. 54, 61–67.
Thimon, L., Peypoux, F., Wallach, J., and Michel, G. (1995). Effect of the lipopeptide antibiotic, iturin A, on morphology and membrane ultrastructure of yeast cells. FEMS Microbiol. Lett. 128, 101–106. doi: 10.1111/j.1574-6968.1995.tb07507.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tijerino, A., Elena, C. R., Moraga, J., Malmierca, M. G., Vicente, F., Aleu, J., et al. (2011). Overexpression of the trichodiene synthase gene tri5 increases trichodermin production and antimicrobial activity in Trichoderma brevicompactum. Fungal Gene. Biol. 48, 285–296. doi: 10.1016/j.fgb.2010.11.012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tobes, R., and Pareja, E. (2005). Repetitive extragenic palindromic sequences in the Pseudomonas syringae pv. tomato DC3000 genome: extragenic signals for genome reannotation. Res. Microbiol. 156, 424–433. doi: 10.1016/j.resmic.2004.10.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tobes, R., and Pareja, E. (2006). Bacterial repetitive extragenic palindromic sequences are DNA targets for insertion sequence elements. BMC Genomics 7:62. doi: 10.1186/1471-2164-7-62
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tsuge, K., Akiyama, T., and Shoda, M. (2001). Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183, 6265–6273. doi: 10.1128/JB.183.21.6265-6273.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
van Rij, E. T., Girard, G., Lugtenberg, B. J., and Bloemberg, G. V. (2005). Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 151, 2805–2814. doi: 10.1099/mic.0.28063-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vetsigian, K., Jajoo, R., and Kishony, R. (2011). Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 9:e1001184. doi: 10.1371/journal.pbio.1001184
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vining, L. C. (1992). Secondary metabolism, inventive evolution and biochemical diversity - a review. Gene 115, 135–140. doi: 10.1016/0378-1119(92)90551-Y
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Viswanathan, R., and Samiyappan, R. (2001). Antifungal activity of chitinases produced by some fluorescent pseudomonads against Colletotrichum falcatum Went causing red rot disease in sugarcane. Microbiol. Res. 155, 309–314. doi: 10.1016/S0944-5013(01)80009-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vizcaino, M. I., Guo, X., and Crawford, J. M. (2014). Merging chemical ecology with bacterial genome mining for secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 41, 285–299. doi: 10.1007/s10295-013-1356-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Voisard, C., Bull, C. T., Keel, C., Laville, J., Maurhofer, M., Schnider, U., et al. (1994). “Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches,” in Molecular Ecology of Rhizosphere Microorganisms, eds F. O'Gara, D. Dowling, and B. Boesten (Weinheim: VCH Publishers), 67–89.
Voisard, C., Keel, C., Haas, D., and Dèfago, G. (1989). Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8, 351–358.
Walker, J., and Abraham, E. (1970). The structure of bacilysin and other products of Bacillus subtilis. Biochem. J. 118, 563–570.
Wallwey, C., and Li, S. (2011). Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat. Prod. Rep. 28, 496–510. doi: 10.1039/C0NP00060D
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, C.-L., Farrand, S. K., and Hwang, I. (1994). Organization and expression of the genes on pAgK84 that encode production of agrocin 84. Mol. Plant Microbe Interact. 7, 472–481. doi: 10.1094/MPMI-7-0472
Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P., and McPhail, A. T. (1971). Plant antitumor agents. VI. isolation and structure of Taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 93, 2325–2327. doi: 10.1021/ja00738a045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wei, C., Hansen, B. S., Vaughan, M. H., and McLaughlin, C. S. (1974). Mechanism of action of the mycotoxin trichodermin, a 12,13-epoxytrichothecene. Proc. Natl. Acad. Sci. U.S.A. 71, 713–717. doi: 10.1073/pnas.71.3.713
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Westman, E. L., Yan, M., Waglechner, N., Koteva, K., and Wright, G. D. (2013). Self resistance to the atypical cationic antimicrobial peptide edeine of Brevibacillus brevis Vm4 by the N-acetyltransferase EdeQ. Chem. Biol. 20, 983–990. doi: 10.1016/j.chembiol.2013.06.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Whatling, C. A., Hodgson, J. E., Burnham, M. K. R., Clarke, N. J., Franklin, F. C. H., and Thomas, C. M. (1995). Identification of a 60 kb region of the chromosome of Pseudomonas fluorescens NCIB 10586 required for the biosynthesis of pseudomonic acid. Microbiology 141, 973–982. doi: 10.1099/13500872-141-4-973
Whitehead, N. A., Barnard, A. M., Slater, H., Simpson, N. J., and Salmond, G. P. (2001). Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25, 365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wissing, F. (1974). Cyanide formation from oxidation of glycine by a Pseudomonas species. J. Bacteriol. 117, 1289–1294.
Xie, N.-Z., Li, J.-X., Song, L.-F., Hou, J.-F., Guo, L., Du, Q.-S., et al. (2014). Genome sequence of type strain Paenibacillus polymyxa DSM 365, a highly efficient producer of optically active (R, R)-2, 3-butanediol. J. Biotechnol. 195, 72–73. doi: 10.1016/j.jbiotec.2014.07.441
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xiong, Z.-Q., Yang, Y.-Y., Zhao, N., and Wang, Y. (2013). Diversity of endophytic fungi and screening of fungal paclitaxel producer from Anglojap yew, Taxus x media. BMC Microbiol. 13:71. doi: 10.1186/1471-2180-13-71
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yanagisawa, T., Lee, J. T., Wu, H. C., and Kawakami, M. (1994). Relationship of protein-structure of isoleucyl-transfer-RNA synthetase with pseudomonic acid resistance of Escherichia coli - proposed mode of action of pseudomonic acid as an inhibitorof isoleucyl-transfer-RNA synthetase. J. Biol. Chem. 269, 24304–24309.
Yang, F., and Cao, Y. (2012). Biosynthesis of phloroglucinol compounds in microorganisms - review. Appl. Microbiol. Biotechnol. 93, 487–495. doi: 10.1007/s00253-011-3712-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yang, K., Han, L., and Vining, L. C. (1995). Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a repressor gene, jadR2. J. Bacteriol. 177, 6111–6117.
Yang, Y., Zhao, H., Barrero, R. A., Zhang, B., Sun, G., Wilson, I. W., et al. (2014). Genome sequencing and analysis of the paclitaxel-producing endophytic fungus Penicillium aurantiogriseum NRRL 62431. BMC Genomics 15:69. doi: 10.1186/1471-2164-15-69
Yazgan, A., Özcengiz, G., and Marahiel, M. A. (2001). Tn10 insertional mutations of Bacillus subtilis that block the biosynthesis of bacilysin. Biochim. Biophys. Acta 1518, 87–94. doi: 10.1016/S0167-4781(01)00182-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zeigler, D. R. (2011). The genome sequence of Bacillus subtilis subsp. spizizenii W23: insights into speciation within the B. subtilis complex and into the history of B. subtilis genetics. Microbiology 157, 2033–2041. doi: 10.1099/mic.0.048520-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, D.-X., Nagabhyru, P., Blankenship, J. D., and Schardl, C. L. (2010). Are loline alkaloid levels regulated in grass endophytes by gene expression or substrate availability? Plant Signal. Behav. 5, 1419–1422. doi: 10.4161/psb.5.11.13395
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, L., Li, X., Zhang, F., and Wang, G. (2014). Genomic analysis of Agrobacterium radiobacter DSM 30147T and emended description of A. radiobacter (Beijerinck and van Delden 1902) Conn 1942 (Approved Lists 1980) emend. Sawada et al. 1993. Stand. Genomic Sci. 9, 574. doi: 10.4056/sigs.4688352
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhao, J., Mou, Y., Shan, T., Li, Y., Zhou, L., Wang, M., et al. (2010). Antimicrobial metabolites from the endophytic fungus Pichia guilliermondii isolated from Paris polyphylla var. yunnanensis. Molecules 15, 7961–7970. doi: 10.3390/molecules15117961
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, R., Rasooly, R., and Linz, J. E. (2000). Isolation and analysis of fluP, a gene associated with hyphal growth and sporulation in Aspergillus parasiticus. Mol. Gen. Genet. 264, 514–520. doi: 10.1007/s004380000335
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, X., Zhu, H., Liu, L., Lin, J., and Tang, K. (2010). A review: recent advances and future prospects of Taxol-producing endophytic fungi. Appl. Microbiol. Biotechnol. 86, 1707–1717. doi: 10.1007/s00253-010-2546-y
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zimmerman, S. B., Schwartz, C. D., Monaghan, R. L., Pelak, B. A., Weissberger, B., Gilfillan, E. C., et al. (1987). Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. I. production, taxonomy and antibacterial activity. J. Antibiot. 40, 1677–1681. doi: 10.7164/antibiotics.40.1677
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zotchev, S. B., Sekurova, O. N., and Katz, L. (2012). Genome-based bioprospecting of microbes for new therapeutics. Curr. Opin. Biotechnol. 23, 941–947. doi: 10.1016/j.copbio.2012.04.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zou, Z., Du, D., Zhang, Y., Zhang, J., Niu, G., and Tan, H. (2014). A γ−butyrolactone−sensing activator/repressor, JadR3, controls a regulatory mini−network for jadomycin biosynthesis. Mol. Microbiol. 94, 490–505. doi: 10.1111/mmi.12752
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: genes, biodiversity, evolution, plant associated microbes, rhizosphere, endophyte, antimicrobial secondary metabolites
Citation: Mousa WK and Raizada MN (2015) Biodiversity of genes encoding anti-microbial traits within plant associated microbes. Front. Plant Sci. 6:231. doi: 10.3389/fpls.2015.00231
Received: 21 January 2015; Accepted: 23 March 2015;
Published: 10 April 2015.
Edited by:
Joanna Marie-France Cross, Inönü University, TurkeyCopyright © 2015 Mousa and Raizada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manish N. Raizada, Department of Plant Agriculture, University of Guelph, 50 Stone Road East, Guelph, ON, N1G 2W1, Canada raizada@uoguelph.ca
 Walaa K. Mousa
Walaa K. Mousa Manish N. Raizada
Manish N. Raizada