- 1Department of Biology, Duke University, Durham, NC, United States
- 2Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 3The Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 4The Bredesen Center for Interdisciplinary Research and Graduate Education, The University of Tennessee, Knoxville, Knoxville, TN, United States
CO2 uptake and water loss in plants are regulated by microscopic pores on the surface of leaves, called stomata. This enablement of gas exchange by the opening and closing of stomata is one of the most essential processes in plant photosynthesis and transpiration, affecting water-use efficiency (WUE) and thus drought susceptibility. In plant species with crassulacean acid metabolism (CAM) photosynthesis, diel stomatal movement pattern is inverted relative to C3 and C4 photosynthesis species, resulting in much higher WUE and drought tolerance. However, little is known about the molecular basis of stomatal movement in CAM species. The goal of this study is to identify candidate genes that could play a role in stomatal movement in an obligate CAM species, Kalanchoë fedtschenkoi. By way of a text-mining approach, proteins were identified in various plant species, spanning C3, C4, and CAM photosynthetic types, which are orthologous to proteins known to be involved in stomatal movement. A comparative analysis of diel time-course gene expression data was performed between K. fedtschenkoi and two C3 species (i.e., Arabidopsis thaliana and Solanum lycopersicum) to identify differential gene expression between the dusk and dawn phases of the 24-h cycle. A rescheduled catalase gene known to be involved in stomatal movement was identified, suggesting a role for H2O2 in CAM-like stomatal movement. Overall, these results provide new insights into the molecular regulation of stomatal movement in CAM plants, facilitating genetic improvement of drought resistance in agricultural crops through manipulation of stomata-related genes.
Introduction
Crassulacean acid metabolism (CAM) is a plant adaptation that involves a carbon concentrating mechanism that is based on a temporal separation of CO2 fixation (Ehleringer and Monson, 1993), which is facilitated by the inverted day/night pattern of stomatal closing and opening in comparison with C3 or C4 photosynthesis species (Males and Griffiths, 2017). Specifically, CAM species open their stomata during the night allowing for uptake of atmospheric CO2, which is converted into malate for storage in the vacuole. During the day, CO2 is released from malate while stomata are closed, resulting in CO2 accumulation around Rubisco for normal photosynthetic processes (Ehleringer and Monson, 1993). Additionally, the inversion of stomatal movement is an important drought avoidance/tolerance mechanism in CAM photosynthesis plants, by which water loss caused by evapotranspiration is decreased and consequently water-use efficiency is increased. Engineering of these traits into non-CAM species has great potential for genetic improvement of drought resistance in crops, which requires a deep understanding of the molecular mechanisms underlying stomatal movement in CAM photosynthesis plants (Borland et al., 2014; Yang et al., 2015).
Aside from stomata research in CAM, research on stomata, in general, has recently increased (Figure 1). In all plants, stomata play essential roles in controlling water losses caused by transpiration and CO2 uptake for photosynthesis, modulating the transpiration-driven water flow through the soil-plant-atmosphere continuum, and plant adaptation to changing environmental conditions and stresses (Damour et al., 2010). Understanding stomatal development, movement, and patterning can facilitate engineering efforts to improve these traits. However, two issues today hinder the pace of advancement in stomatal research. Firstly, most work on these stomatal processes has been conducted in the model C3 plant Arabidopsis thaliana. This can present an issue when wanting to transfer the knowledge gained from these studies to another species as the genes involved in these processes can vary in regulation and/or function, as seen in CAM species (Abraham et al., 2016; Males and Griffiths, 2017; Yang et al., 2017), and in the grass species Brachypodium distachyon (Raissig et al., 2016, 2017). Secondly, homology between protein sequences can help infer protein function (Clark and Radivojac, 2011), but the issue is that many proteins’ functional information is missing from annotation databases or is hidden in the scientific literature. For instance, a protein of interest could be homologous to a protein characterized to function in a stomata-related process, but this information is not known because the characterized protein lacks the gene ontology (GO) annotation.
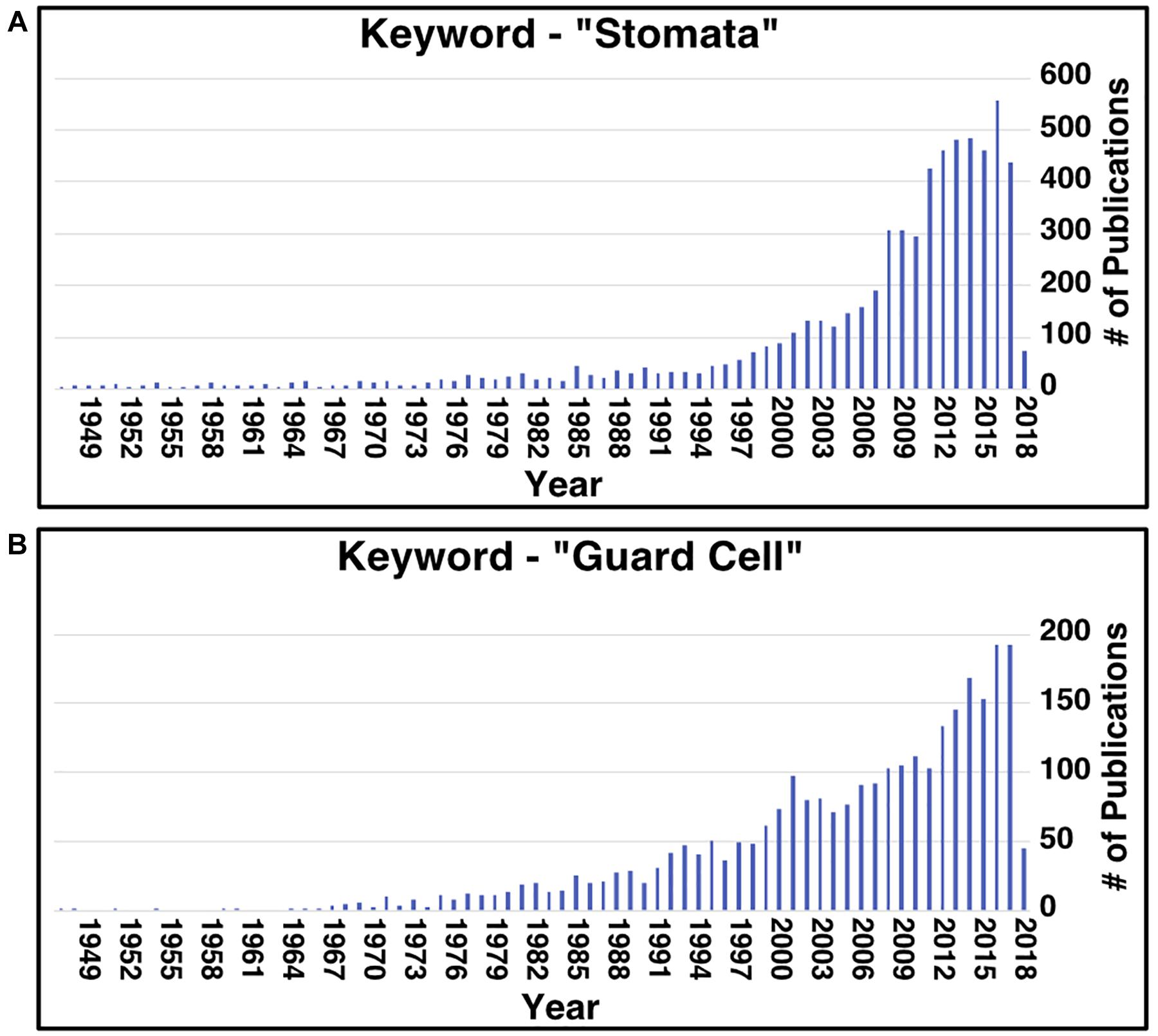
Figure 1. Number of publications per year related to stomata and guard cell research since 1949. Results of searching the PubMed database using the key words “stomata” (A) and “guard cell” (B).
There are text-mining tools that can be used to search for information about similar proteins by combining BLAST searches with links to articles from certain databases (Gilchrist et al., 2008; Li et al., 2013; Jaroszewski et al., 2014). However, these tools do not search the literature, therefore, limiting their analysis. Recently, a new web-based tool called PaperBLAST (Price and Arkin, 2017), was developed to combine sequence-homology searches and text-mining of linked articles for predicting a protein’s function. PaperBLAST has potential to alleviate the two issues facing stomatal research described above, and therefore aid in discovering molecular mechanisms behind inverted stomatal movement seen in CAM species.
In this study, we first identified underexplored stomatal-related genes that contain no stomata-related annotations but have been linked to stomatal-related processes using PaperBLAST. Next, we identified genes that could be involved in the inverted stomatal movement in CAM plants through a comparative analysis of time-course gene expression data between an obligate CAM photosynthesis species (Kalanchoë fedtschenkoi) and two C3 photosynthesis species (A. thaliana and Solanum lycopersicum). Our research greatly expanded the catalog of stomata-related genes in plants, providing a resource for future experimental work in stomatal research of non-model species, and more importantly, providing gene targets for engineering CAM-like traits into non-CAM species.
Materials and Methods
Ortholog Groups and Phylogenetic Analysis
Ortholog groups consisting of proteins from 26 plant species were obtained from Yang et al. (2017). For phylogenetic analysis, protein sequence alignments were created using the web-based MUSCLE program1 with default parameters (Katoh and Standley, 2013; McWilliam et al., 2013; Li et al., 2015). Phylogenetic trees were generated from the protein sequence alignments using the web-based IQ-Tree program2 with default parameters (Trifinopoulos et al., 2016).
Gene Ontology Analysis of Protein Sequences
Gene ontology terms for A. thaliana and K. fedtschenkoi genes were taken from the gene annotation information downloaded from Phytozome v12.13 (Goodstein et al., 2012). The right-sided hypergeometric enrichment test at medium network specificity in ClueGO (Bindea et al., 2009) was used to identify the biological processes over-represented in individual gene sets. The Holm-Bonferroni step-down method (Holm, 1979) was performed for p-value correction. The minimum and maximum thresholds for selected GO-tree levels were 3 and 8, respectively, while individual clusters were required to include no less than 3% of genes associated with each GO term. To minimize GO-term redundancy, GO-term fusion and grouping settings were selected and the representative term for each functional cluster was determined as the term enriched at the highest level of significance. The GO terms with p-values less than or equal to 0.05 were considered significantly enriched.
Data-Mining Using PaperBLAST
To include a variety of evolutionary lineages, the proteomes of 13 representative plant species including Amborella trichopoda (basal angiosperm, C3 photosynthesis), Ananas comosus (monocot, CAM photosynthesis), A. thaliana (dicot, C3 photosynthesis), B. distachyon (monocot, C3 photosynthesis), K. fedtschenkoi (dicot, CAM photosynthesis), Mimulus guttatus (dicot, C3 photosynthesis), Musa acuminata (monocot, C3 photosynthesis), Oryza sativa (monocot, C3 photosynthesis), Phalaenopsis equestris (monocot, CAM photosynthesis), Setaria italica (monocot, C4 photosynthesis), S. lycopersicum (dicot, C3 photosynthesis), Sorghum bicolor (monocot, C4 photosynthesis), and Vitis vinifera (dicot, C3 photosynthesis) were downloaded from the PLAZA 4.0 database (Van Bel et al., 2017; Supplementary Table S1). Each protein sequence was used as a search query in PaperBLAST (Price and Arkin, 2017) using an E-value threshold of 1e-3. The subsequent HTML files contain various information on the homologous proteins, such as functional information, article name, and text snippets from articles containing the homologous protein’s IDs. The HTML files were collected and parsed through searching for any presence of stomata-related keywords (i.e., “stomata” and “guard cell”) in each PaperBLAST hit’s affiliated data using in-house python scripts. Any homologous proteins that did not contain the keywords in their related data were filtered out. The relevant extracted data for each species are summarized in Supplementary Table S2.
Analysis of Time-Course Gene Expression Data
The diel expression data for K. fedtschenkoi were taken from Yang et al. (2017), which were generated from transcriptome-sequencing (RNA-Seq) of whole leaf tissue samples collected at 2-h intervals from plants grown under a 12-h light/12-h dark cycle. The diel expression data for A. thaliana were taken from Mockler et al. (2007), which were generated from samples collected at 4-h intervals from plants grown under a 12-h light/12-h dark cycle. Raw RNA-Seq reads for S. lycopersicum were taken from the DDBJ Sequence Read Archive4 under the accession numbers DRA003529 and DRA0035530, which were generated from whole-leaf samples collected every 2 h from plants grown in a 10-h light/14-h dark cycle (Higashi et al., 2016). The S. lycopersicum raw sequencing reads were quality checked by FastQC (Andrew, 2014), trimmed using Trimmomatic v0.36 (Bogler et al., 2014) with default parameters, and quality checked again by FastQC. The trimmed S. lycopersicum RNA-Seq reads were mapped to the S. lycopersicum iTAG v2.3 reference genome using TopHat2 v1.0.1 (Kim et al., 2013) using default parameters. Read counts were computed using Cufflink v2.2.1 (Trapnell et al., 2012) and fragments per kilobase of transcript per million mapped reads (FPKM) were calculated. Processing of all raw sequencing data was performed using Kbase (Arkin et al., 2018). Genes with an average FPKM less than 0.01 and with less than half of their sampling points having an FPKM equal to zero were filtered out from the K. fedtschenkoi and S. lycopersicum expression data sets. Following previous works utilizing the A. thaliana time course transcriptome dataset (Abraham et al., 2016; Yang et al., 2017; Moseley et al., 2018), the A. thaliana data was adjusted to arrive at expression profiles on the same time scale as K. fedtschenkoi and S. lycopersicum gene expression data. Specifically, the cubic interpolation algorithm in the pandas Python library5 was used to simulate the A. thaliana gene expression data at additional time points so that all three data sets consisted of sampling points spaced every 2 h over a 24-h period.
Comparative Analysis of Gene Expression
Comparative analysis of transcript expression between the time-windows dusk and dawn was performed as described by Yang et al. (2017). For both K. fedtschenkoi and A. thaliana, the dusk-window covers 10, 12, and 14 h after the starting of the light period and the dawn-window covers 22, 24, and 2 h after the starting of the light period (Supplementary Figure S1). For S. lycopersicum, which was grown under a slightly different light regime than K. fedtschenkoi and A. thaliana, the dusk-window covers 8, 10, and 12 after the starting of the light period, and the dawn-window covers 22, 24, and 2 h after the starting of the light period (Supplementary Figure S1). For each species, any genes with negative gene expression levels were removed and expression data for each gene was normalized as described in Yang et al. (2017). A two-column matrix was created, where rows represented genes, one column represented the sum of all transformed dusk time points, and the other column represented the sum of all transformed dawn time points. For each gene, the right-tailed Fisher Exact Test was used to determine if that gene’s expression was enriched in dusk, dawn, or neither, according to the contingency tables described in Yang et al. 2017; (Supplementary Figure S1). The Fisher Exact Test was performed using the fisher_exact function from the scipy Python library6. The False Discovery Rate (Benjamini and Hochberg, 1995) was controlled per species and time-window at a p-value of < 0.05. This comparative analysis was performed to identify ortholog groups containing re-scheduled gene expression in CAM species in comparison with C3 species, in which the S. lycopersicum and A. thaliana gene were enriched in the same time-window whereas the K. fedtschenkoi gene was enriched in the opposite time-window (i.e., dawn vs. dusk).
Results
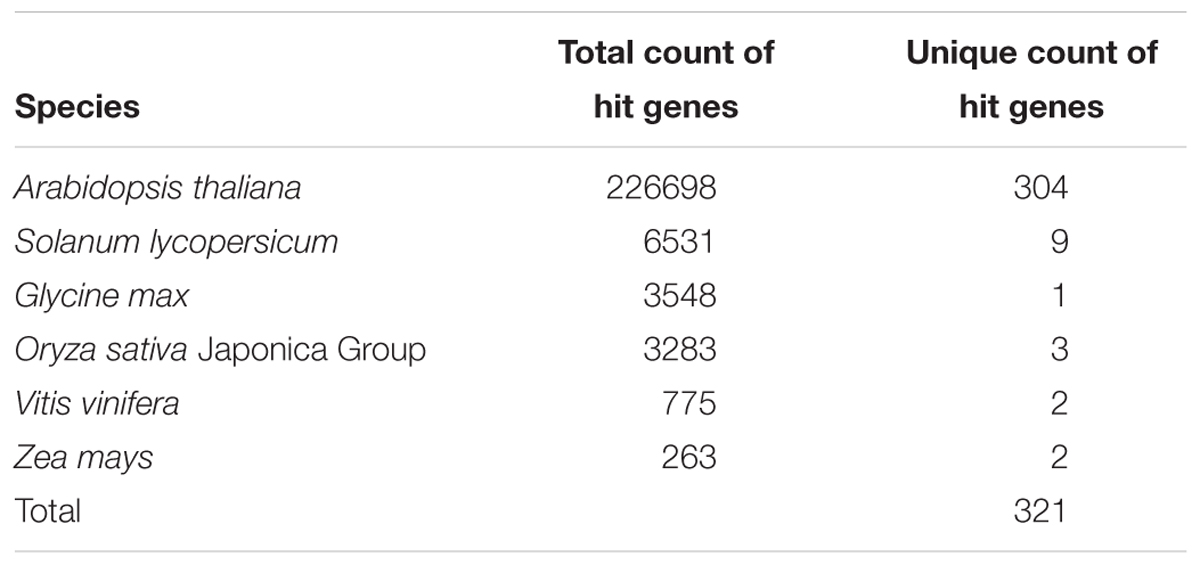
Identification of Stomata-Related Genes
Our PaperBLAST analysis of the protein sequences in 13 plants species identified an average of 5,196 proteins per species that could be involved in stomata-related processes, accounting for 16.4% of the respective plant genome on average (Supplementary Table S3). A total of 321 proteins in the PaperBLAST database had data containing the keywords “stomata” and “guard cell” (Table 1). Most of these proteins (304) were A. thaliana proteins, with the second most (9) belonging to S. lycopersicum. Only A. thaliana and S. lycopersicum had proteins that matched to all 321 proteins in the PaperBLAST database (Supplementary Table S3). Each protein in PaperBLAST is linked to a publication(s) that contains information on the respective protein. 272 publications were cataloged containing at least one of the 321 proteins (Supplementary Table S3). Since a majority of the 321 proteins were A. thaliana proteins and that A. thaliana is a well-annotated organism, annotations of the 304 A. thaliana proteins were examined. Using the current annotation of A. thaliana proteins, 178 proteins were annotated with stomata- or guard cell-related terms. An additional 20 genes known to play key roles in stomatal movement and development were added as well (Ohashi-Ito and Bergmann, 2006; MacAllister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008; Kollist et al., 2014), totaling 198 stomata-related proteins with functional annotation either GO or are known as key stomatal-related genes (Supplementary Table S4). Among the 304 A. thaliana stomata-related proteins in the PaperBLAST database, only 97 proteins were annotated as stomata-related in the GO database or described as key stomatal-related genes, indicating that there are 207 stomata-related genes missed by GO and hidden in the scientific literature (Supplementary Table S5 and Figure 2A). These 207 genes will be referred to as underexplored stomata-related genes/proteins from here on. The biological processes overrepresented in these 207 underexplored A. thaliana stomata-related proteins include signal transduction, phosphorylation-related processes, and several response processes (Figure 2B).
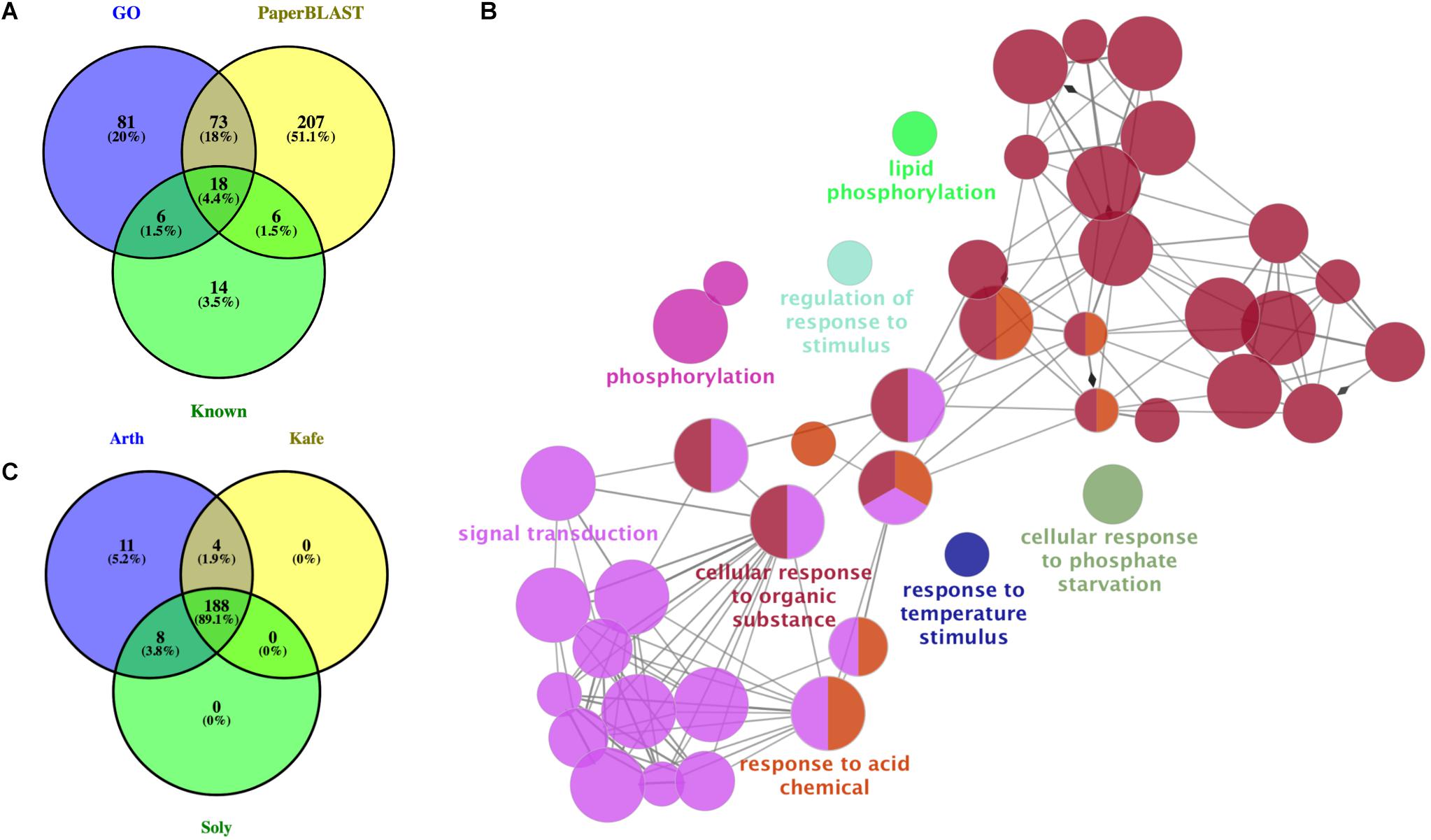
Figure 2. Assessment and identification of new stomata-related genes. (A) Overlap of Arabidopsis thaliana genes with annotations related to stomata, known as key stomatal genes, and identified in PaperBLAST database as stomata-related. GO: genes annotated as stomata-related from Gene Ontology analysis. Known: genes known as stomata-related from the literature. PaperBLAST: stomata-related genes identified by using PaperBLAST search. (B) Gene ontology enrichment of the 207 new A. thaliana stomatal genes. (C) Overlap of ortholog groups between new stomata-related genes in A. thaliana (Arth), Solanum lycopersicum (Soly), and Kalanchoë fedtschenkoi (Kafe).
PaperBLAST uses an E-value threshold of 1e-3 to infer homology between two proteins, and generally, this E-value cutoff can be used reliably for this purpose (Pearson, 2013). However, to produce a more confident set of stomata-related genes for each species, the ortholog groups (OGs) of the 321 proteins linked to stomata-related processes in PaperBLAST were extracted. In the construction of these OGs, Yang et al. (2017) used a more stringent E-value threshold of 1e-5 to infer homology and included proteins from 26 plant species. A total of 257 OGs were extracted for the 321 proteins linked to stomata-related processes in PaperBLAST. Individually, the number of proteins from each species that shared the same OG as the PaperBLAST protein ranged from just over 500 proteins in A. trichopoda to over 1,200 proteins in both A. thaliana and Musa acuminata (Supplementary Table S6). Further categorization of these OGs on whether the respective species’ proteins were orthologous to an A. thaliana protein in any of the three stomata gene categories (i.e., annotated as stomata- /guard cell-related, known key stomatal genes, or underexplored stomata-related genes) is shown in Supplementary Table S7.
Rescheduled Diel Expression of Stomatal Genes in K. fedtschenkoi
It is hypothesized that the rewiring of diel gene expression pattern played a role in the evolution of CAM plants from C3 plants (Yang et al., 2017). To determine if any of the K. fedtschenkoi genes orthologous to an A. thaliana gene in any of the three stomata gene categories could play a role in the inversion of day/night stomatal movement pattern, their diel expression profiles were compared to their ortholog’s expression profiles in S. lycopersicum and A. thaliana. Specifically, the gene expression profiles of 188 OGs, which were selected as they contained stomata-related genes identified from the previous section in K. fedtschenkoi, S. lycopersicum, and A. thaliana (Figure 2C).
Two criteria were used to identify stomata-related genes that display rescheduled (dawn vs. dusk) gene expression in the CAM photosynthesis species (K. fedtschenkoi) in comparison with the two C3 photosynthesis species (S. lycopersicum and A. thaliana): (1) The K. fedtschenkoi gene must be significantly enriched in either dawn or dusk, whereas the S. lycopersicum and A. thaliana orthologs must be significantly enriched in the opposite time-window (i.e., dawn vs. dusk), and (2) the K. fedtschenkoi gene must have a spearman rank correlation coefficient < -0.6 with both the S. lycopersicum and A. thaliana orthologs, whereas the S. lycopersicum and A. thaliana orthologs must have a Spearman rank correlation coefficient > 0.6 among themselves. Based on these two criteria, 16 OGs were identified to contain stomata-related genes showing rescheduled (dawn vs. dusk) gene expression in the CAM photosynthesis species in comparison with the two C3 photosynthesis species (Table 2 and Supplementary Figure S2). Among these 16 stomata-related OGs, only two OGs contained A. thaliana genes annotated as stomata-related genes: CIPK23 (AT1G30270) and RCAR3 (AT5G53160) (Table 2). The remaining 14 OGs contained A. thaliana genes that are not annotated or known as key stomata-related genes but have been reported to be involved in a stomata-related process in the literature, or in other words, underexplored stomata-related genes.
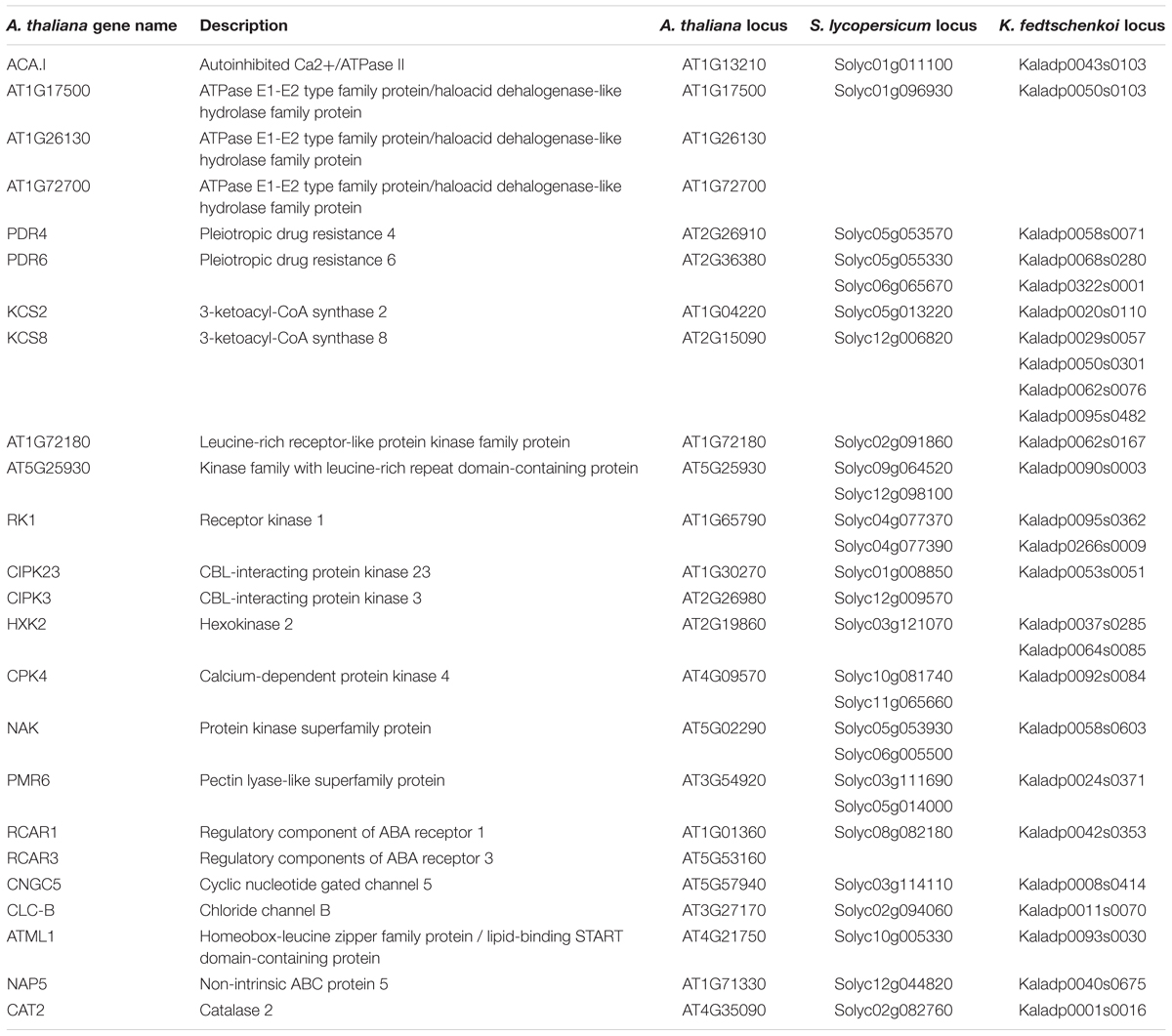
Table 2. Orthologous genes in Arabidopsis thaliana, Solanum lycopersicum, and Kalanchoë fedtschenkoi that displayed differential enrichment between dusk and dawn in gene expression.
Evolutionary Dynamics of Stomata-Related Genes
The three plant species studied here have undergone multiple rounds of whole-genome duplication (Yang et al., 2017), which can cause multiple copies (i.e., paralogs) of one gene. At large, gene duplications have contributed to the evolution of novel functions (e.g., adaptation to stress) via the subsequent dynamic events that can occur after gene duplication, such as subfunctionalization and neofunctionalization (Yang et al., 2006; Panchy et al., 2016). To better understand the evolutionary dynamics that supported CAM arising from C3, the phylogenetic relationships within the 16 OGs were examined. Using A. trichopoda, which is a basal angiosperm plant species (Albert et al., 2013), as an outgroup, phylogenetic trees were constructed for each OG.
The OG containing the A. thaliana RCAR3 gene also included two A. thaliana paralogs identified as RCAR1 (AT1G01360) and RCAR2 (AT4G01026). Only two genes from K. fedtschenkoi and S. lycopersicum each were placed in this OG. Two subclades were identified in this OG, one containing the A. thaliana paralogs RCAR1 and RCAR2 and a S. lycopersicum gene (SL08G082180) and the other subclade containing the A. thaliana RCAR3 gene, a S. lycopersicum gene (SL03G007310), and the two K. fedtschenkoi genes (Kaladp0008s0082 and Kaladp0042s0353) (Supplementary Figure S3). RCAR genes are known to function as ABA receptors and mediate ABA-dependent regulation of type 2C protein phosphatases (Ma et al., 2009). In the presence of ABA, RCAR genes inhibit type 2C protein phosphatases phosphatase activities, which allows SNF1-related protein kinase 2 protein kinases to activate components involved in regulating stomatal movement (Culter et al., 2010; Raghavendra et al., 2010). The K. fedtschenkoi RCAR3 gene was identified as having rescheduled gene expression relative to the A. thaliana RCAR3 but not the S. lycopersicum RCAR3 (Supplementary Figure S3). Previous work examining the temporal differences in gene expression between the CAM species Agave Americana and A. thaliana found similar evidence of rescheduled gene expression between RCAR3 orthologs (Abraham et al., 2016). Determining the specific roles of each RCAR gene in ABA signaling, particularly in CAM species, is needed to better understand the differences in their gene expression between CAM and C3 species and the impact the differences could have on stomatal movement.
The phylogenetic tree of the OG containing the A. thaliana powdery mildew resistant 6 (PMR6, AT3G54920) gene displayed rich evolutionary dynamics as multiple genes from each of the three species were found within this OG (Supplementary Figure S4). The rescheduled genes identified in this study were not placed near each other in an evolutionary sense as the S. lycopersicum genes (SL03G111690, SL05G014000) were found several branching events away from the A. thaliana (AT3G54920) and K. fedtschenkoi (Kaladp0024s0371) genes. However, the A. thaliana and K. fedtschenkoi genes were closely related to each other, as well as to one A. thaliana (AT5G04310) and K. fedtschenkoi (Kaladp0008s0911) gene each and three S. lycopersicum genes (SL03G071570, SL05G055510, and SL11G008140). Kaladp0024s0371, identified as rescheduled relative to the A. thaliana PMR6 gene, is predominantly expressed during the day, while the A. thaliana PMR6 gene is predominantly expressed toward the end of night time (Supplementary Figure S4). The remaining closely related genes displayed similar expression covering approximately the same time window as the A. thaliana PMR6 gene. PMR6 encodes a pectate lyase-like protein and mutations in PMR6 have shown to alter the composition of the plant cell wall via increases in pectin (Vogel, 2002). Moreover, stomata open more in pmr6 mutants than in wild-type likely due to the increased cell wall stiffness of the guard cell (Woolfenden et al., 2017). Whether alteration of cell wall composition during the day contributed to the inversion of stomatal movement seen in CAM plants cannot be determined here. Further investigation into the temporal dynamics of cell wall composition in the guard cells of CAM plants would provide better clues to the role of PMR6 in stomatal movement, and to guard cell physiology in CAM plants as a whole.
Two OGs containing underexplored genes had outgroup rooted trees confidently constructed, with one OG containing protein kinases and the other containing catalases. The protein kinase OG contained two A. thaliana genes reported to be involved in light-activation of stomatal opening, APK1a (AT1G07570) and APK1b (AT2G28930) (Elhaddad et al., 2014) and a third reported to be involved in ABA-induced osmotic stress response, NAK (AT5G02290) (Kodama et al., 2009). The A. thaliana NAK gene formed a subclade with three S. lycopersicum genes and two K. fedtschenkoi genes (Figure 3A), potentially representing duplication events after divergence from a common ancestor. The K. fedtschenkoi NAK gene, Kaladp0058s0603 (NAK1), was found to have rescheduled expression in comparison with the A. thaliana NAK gene, and one of the S. lycopersicum NAK genes (SL06G010850). The remaining NAK genes in S. lycopersicum and K. fedtschenkoi displayed expression covering approximately the same time window as their respective paralogs. Little information is available on the functional role of NAK, but Kodama et al. (2009) have reported that under drought conditions, ABA induces NAK gene expression. NAK autophosphorylates and migrates into the nucleus and phosphorylates subsequent proteins. They propose this results in modulation of nuclear function to cope with dehydration stress. K. fedtschenkoi NAK1 peaks in the morning, while the NAK genes in the C3 plants are expressed during dusk. CAM arose as an adaptation to water-limited environments, so the rescheduled expression of an osmotic stress response gene, which is closely related to genes involved in stomatal opening, represents an interesting mode of CAM evolution.
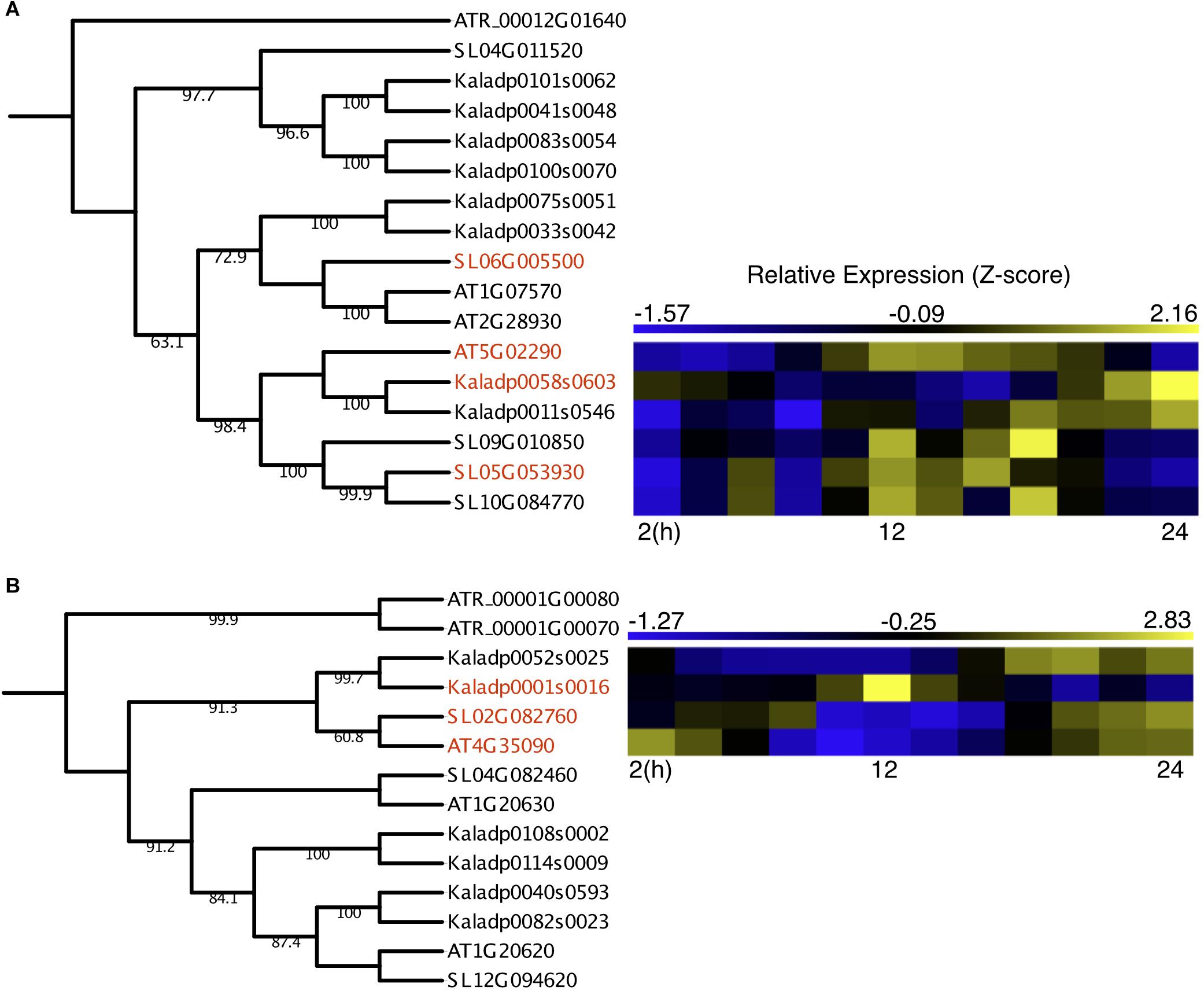
Figure 3. Rescheduled diel expression of stomata-related genes in a CAM photosynthesis species (K. fedtschenkoi) in comparison with two C3 photosynthesis species (A. thaliana and S. lycopersicum). (A) Phylogenetic tree of protein kinase genes and expression heatmaps of NAK genes in A. thaliana, S. lycopersicum, and K. fedtschenkoi. (B) Phylogenetic tree of catalase genes and expression heatmaps of CAT2 genes in A. thaliana, S. lycopersicum, and K. fedtschenkoi. Gene names in red text indicate genes found to be enriched in either dawn or dusk.
The catalase OG contained three genes in each of the two C3 photosynthesis species (A. thaliana and S. lycopersicum), consistent with a previous study (Mhamdi et al., 2012). However, there were 6 K. fedtschenkoi genes in the catalase OG, which may result from a recent whole-genome duplication in K. fedtschenkoi (Yang et al., 2017). The A. thaliana gene in the catalase OG highlighted in Figure 3B, which contains rescheduled gene expression in the CAM species in comparison with the C3 species, is annotated as catalase 2 (CAT2) (Table 2). CAT2 is a part of the photorespiratory pathway and aids in the detoxification of H2O2 (Queval et al., 2007; Mhamdi et al., 2010, 2012). The CAT2 clade in the catalase OG phylogenetic tree (Figure 3B) includes all the genes listed in Table 2 for this OG but also includes another K. fedtschenkoi gene (Kaladp0052s0025), suggesting there are two CAT2 genes in K. fedtschenkoi. The A. thaliana and S. lycopersicum genes have gene expression enriched and peaking in the morning, as well as one of the K. fedtschenkoi CAT2 (CAT2.1) genes (Figure 3B). The second K. fedtschenkoi gene (CAT2.2) has gene expression enriched and peaking during dusk (Figure 3B). This presents an interesting scenario as the proteins that each catalase gene encodes for, have relatively specific roles in determining the accumulation of H2O2 produced through various metabolic pathways (Mhamdi et al., 2012).
Discussion
Research into stomata’s role in controlling water loss and gas exchange as well as in facilitating protection against changing environmental conditions and stresses has increased recently, with the purpose of understanding the molecular mechanisms behind these features. However, the functional information generated from these studies is either missing from annotation databases or is hidden in the scientific literature (Price and Arkin, 2017), therefore, hindering the progress in functional genomics research to gain a deep and comprehensive understanding of stomata-related genes. To help alleviate this problem, this study identified genes that were neither annotated nor known as key stomata-related genes across 13 plant species, but have been reported as being involved in a stomata-related process in the literature. To demonstrate the utility of this new resource for stomata-related genes, the molecular mechanism behind the inversion of stomatal movement in CAM species, relative to C3 species, was investigated using the gene sets generated for one CAM species (K. fedtschenkoi) and two C3 species (A. thaliana and S. lycopersicum). Several genes were identified as candidates for further investigation into inverted stomatal movement in CAM.
A little over 300 genes were found in the literature to be involved in stomata-related processes, with a majority of them belonging to A. thaliana. Stomata-related genes were identified in each of the 13 plant species used in this study using PaperBLAST. The subset of genes for each species were further categorized based on sequence similarity with A. thaliana genes found with PaperBLAST, which were either previously annotated/known stomata-related genes, or underexplored stomata-related genes. The underexplored stomata-related genes reported in this study would serve as an excellent resource for future investigations into the molecular mechanisms behind stomata-related processes, particularly in non-Arabidopsis species. The genes identified in the 12 plant species other than A. thaliana could have other functional roles, even though they belong in the same ortholog group. For instance, it has been recently reported that orthologous genes involved in stomatal development and patterning in B. distachyon and A. thaliana display divergence in stomata-related function and regulation, even though they are orthologous genes (Raissig et al., 2016, 2017).
To investigate the molecular mechanism(s) that could help explain the inversion of stomatal movement seen in CAM plants, relative to C3 plants, the newly generated list of homologous stomata-related genes across 13 plant species was examined. CAM plants close their stomata during the day to reduce their rate of transpiration, thus enabling them to better tolerate drought-stress than C3 plants. ABA plays an essential role in a plant’s response to drought by facilitating stomatal closure (Vishwakarma et al., 2017), and in CAM plants, ABA concentrations have been altered to peak right before morning (Abraham et al., 2016). The NAK gene has been characterized to be induced by ABA in drought conditions (Kodama et al., 2009) and was identified in the list of homologous stomatal-related genes as having rescheduled gene expression between K. fedtschenkoi and two C3 species. Assuming the NAK1 gene in K. fedtschenkoi has the same function as its A. thaliana ortholog, it can be hypothesized that the shift in its expression could likely result from or result in the change in ABA concentrations. This hypothesis can be tested using molecular genetics and functional approaches in the future (e.g., characterization of knockout nak1 mutants in K. fedtschenkoi). Determining the role of NAK1 in K. fedtschenkoi, and how it relates to stomatal movement, could provide new insights into the molecular mechanisms which CAM plants use to adapt to a dry environment. Moreover, identifying the regulator of NAK1 could also help in understanding ABA signaling, and therefore stomatal closing, in CAM plants.
An additional gene identified in the list of homologous stomatal-related genes as having undergone rescheduling of gene expression in K. fedtschenkoi relative to C3 species was a catalase gene, CAT2. There are generally three catalase genes in a plant’s genome, and each gene is considered functionally conserved between species (Mhamdi et al., 2012). Specific to the CAT2 gene, CAT2 is expressed in photosynthetic mesophyll cells and guard cells, is involved in photorespiration, and shows day-night rhythms in transcript abundance, with peaks in the morning (Zhong et al., 1994; Zhong and McClung, 1996; Queval et al., 2007; Mhamdi et al., 2010; Mhamdi et al., 2012). CAT2’s primary role is in H2O2 detoxification, specifically H2O2 produced as a result of photosynthesis (Mhamdi et al., 2010). Rubisco is a bifunctional enzyme that also catalyzes oxygenation of RuBP, which produces 2-phosphogylycolate as one of its products. This small molecule is not metabolized via the Calvin-Benson cycle but is dephosphorylated to produce glycolate, which is transported to the peroxisomes. Within the peroxisomes, glycolate is oxidized to glyoxylate using oxygen as a co-factor, which results in H2O2 being produced. This production is negatively controlled by CAT2 (Figure 4A).
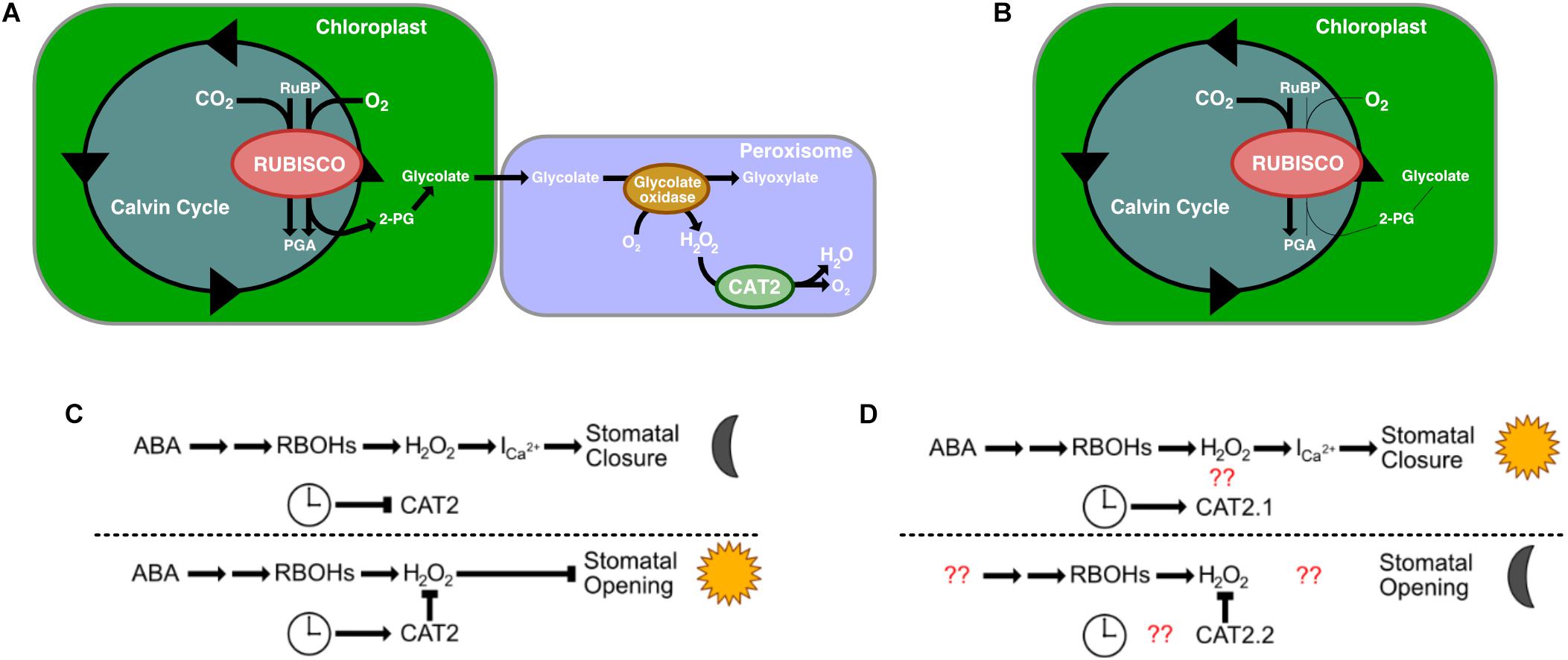
Figure 4. The role of Catalase 2′s (CAT2) roles in C3 plants and in CAM plants. (A) CAT2’s role in photorespiration in C3 plants. (B) Photorespiration is reduced in CAM plants. (C) CAT2’s role in stomatal open closing (top) and opening (bottom) in C3 plants. (D) CAT2’s predicted role in stomatal closing (top) and opening (bottom). Red question marks indict unknown interactions that warrant further investigation. RuBP, Ribulose 1,5-biphosphate; PGA, 3-phosphoglyceric acid; 2-PG, 2-phosphogylycolate; Ica2+, Ca2+ channels; H2O2, hydrogen peroxide; RBOH, respiratory burst oxidases.
CAM is a mechanism to reduce photorespiration by reducing the CO2:O2 ratio in cells (Figure 4B). In other organisms that have evolved a method to reduce glycolate production, the main fate of glycolate is excretion or oxidation to glyoxylate via a mitochondrial dehydrogenase using NAD+ as the final electron acceptor (Stabenau and Winkler, 2005). Whether this is the case in K. fedtschenkoi is yet to be determined, however, our results suggest a reduced role for CAT2.1 in K. fedtschenkoi. Interestingly, CAT2 seems to be duplicated in K. fedtschenkoi, with one copy having conserved gene expression and the other copy being expressed at dawn (Figure 3B). In a study in A. thaliana, CAT2 was found to be involved in ABA signaling for stomatal closure as cat2 mutants had significantly enhanced ABA-induced stomatal closure (Jannat et al., 2011). These authors presented a model where ABA activates respiratory burst oxidases (RBOHs) in the membranes of guard cells which results in rapid production of H2O2 in the guard cells (Kwak et al., 2003). The H2O2 then activates Ca2+ channels causing stomatal closure during the night (Pei et al., 2000; Murata et al., 2001; Figure 4C). CAT2 plays a negative role in ABA signaling in stomatal closure by detoxifying the guard cells. This results in Ca2+ channels not being activated which leaves the stomata open (Figure 4C). Due to the role of CAT2 in stomatal opening, we propose that CAT2.2, expressed during dusk in K. fedtschenkoi, could play a role in stomatal opening by inhibiting ABA-induced stomatal closure by reducing H2O2 concentrations (Figure 4D). Several other authors have suggested that H2O2 activation of Ca2+ channels represents a possible convergent point for multiple stress signaling pathways (Gechev and Hille, 2005; Sewelam et al., 2014; Niu and Liao, 2016). Functional characterization of both CAT2 genes in K. fedtschenkoi is needed to determine any roles in stomatal opening, as well as determine if CAT2.2 is circadian regulated. Further determination of the role of H2O2 signaling in K. fedtschenkoi stomatal movement is needed.
In summary, this study cataloged hundreds of underexplored stomata-related genes in multiple plant species including C3, C4, and CAM photosynthesis plants. We also identified numerous underexplored stomata-related genes that displayed re-scheduled gene expression between orthologs in a CAM species and two C3 species, providing valuable candidates for CAM-engineering in C3 photosynthesis crops to enhance drought-resistance. Furthermore, the impact of gene duplication and diversification on CAM evolution was underlined, highlighting the evolutionary dynamics involved in CAM evolution.
Author Contributions
RM and XY conceived the research. RM performed all data analyses and wrote the manuscript. GT and XY provided input during the study and edited the manuscript.
Funding
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the United States Department of Energy. This research was supported by the United States Department of Energy, Office of Science, Genomic Science Program under award number DE-SC0008834 and the Center for Bioenergy Innovation. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the United States Department of Energy under Contract Number DE-AC05-00OR22725.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank David Weston, Margaret E. Staton, and Jay Chen for their critical comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00292/full#supplementary-material
Footnotes
- ^https://www.ebi.ac.uk/Tools/msa/muscle/
- ^http://www.iqtree.org/
- ^https://phytozome.jgi.doe.gov/pz/portal.html
- ^https://www.ddbj.nig.ac.jp/index-e.html
- ^https://pandas.pydata.org/
- ^https://www.scipy.org/
References
Abraham, P. E., Yin, H., Borland, A. M., Weighill, D., Lim, S. D., De Paoli, H. C., et al. (2016). Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat. Plants 2:16178. doi: 10.1038/nplants.2016.178
Albert, V. A., Barbazuk, W. B., dePamphilis, C. W., Der, J. P., Leebens-Mack, J., Ma, H., et al. (2013). the amborella genome and the evolution of flowering plants. Science 342, 1241089–1241089. doi: 10.1126/science.1241089
Andrew, S. (2014). FastQC A Quality Control tool for High Throughput Sequence Data. Berlin: ScienceOpen.
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., et al. (2018). KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569. doi: 10.1038/nbt.4163
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
Bogler, A., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borland, A. M., Hartwell, J., Weston, D. J., Schlauch, K. A., Tschaplinski, T. J., Tuskan, G. A., et al. (2014). Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci. 19, 327–338. doi: 10.1016/j.tplants.2014.01.006
Clark, W., and Radivojac, P. (2011). Analysis of protein function and its prediction from amino acid sequence. Proteins 79, 2086–2096. doi: 10.1002/prot.23029
Culter, S., Rodriquez, P., Finkelstein, R., and Abrams, S. (2010). Absicsic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Damour, G., Simonneau, T., Cochard, H., and Urban, L. (2010). An overview of models of stomatal conductance at the leaf level. Plant Cell Environ. 33, 1419–1438. doi: 10.1111/j.1365-3040.2010.02181.x
Ehleringer, J. R., and Monson, R. K. (1993). Evolutionary and ecological aspects of photosynthetic pathway variation. Annu. Rev. Ecol. Syst. 24, 411–439. doi: 10.1146/annurev.es.24.110193.002211
Elhaddad, N. S., Hunt, L., Sloan, J., and Gray, J. E. (2014). Light-induced stomatal opening is affected by the guard cell protein kinase APK1b. PLoS One 9:e97161. doi: 10.1371/journal.pone.0097161
Gechev, T. S., and Hille, J. (2005). Hydrogen peroxide as a signal controlling plant programmed cell death. J. Cell Biol. 168, 17–20. doi: 10.1083/jcb.200409170
Gilchrist, M. J., Christensen, M. B., Harland, R., Pollet, N., Smith, J. C., Ueno, N., et al. (2008). Evading the annotation bottleneck: using sequence similarity to search non-sequence gene data. BMC Bioinformatics 9:442. doi: 10.1186/1471-2105-9-442
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Higashi, T., Tanigaki, Y., Takayama, K., Nagano, A. J., Honjo, M. N., and Fukuda, H. (2016). Detection of diurnal variation of tomato transcriptome through the molecular timetable method in a sunlight-type plant factory. Front. Plant Sci. 7:87. doi: 10.3389/fpls.2016.00087
Jannat, R., Uraji, M., Morofuji, M., Hossain, M. A., Islam, M. M., Nakamura, Y., et al. (2011). The roles of CATALASE2 in abscisic acid signaling in Arabidopsis guard cells. Biosci. Biotechnol. Biochem. 75, 2034–2036. doi: 10.1271/bbb.110344
Jaroszewski, L., Koska, L., Sedova, M., and Godzik, A. (2014). PubServer: literature searches by homology. Nucleic Acids Res. 42, W430–W435. doi: 10.1093/nar/gku450
Kanaoka, M. M., Pillitteri, L. J., Fujii, H., Yoshida, Y., Bogenschutz, N. L., Takabayashi, J., et al. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20, 1775–1785. doi: 10.1105/tpc.108.060848
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, 1–13. doi: 10.1186/gb-2013-14-4-r36
Kodama, Y., Tamura, T., Hirasawa, W., Nakamura, K., and Sano, H. (2009). A novel protein phosphorylation pathway involved in osmotic-stress response in tobacco plants. Biochimie 91, 533–539. doi: 10.1016/j.biochi.2009.01.003
Kollist, H., Nuhkat, M., and Roelfsema, M. R. (2014). Closing gaps: linking elements that control stomatal movement. New Phytol. 203, 44–62. doi: 10.1111/nph.12832
Kwak, J. M., Mori, I. C., Pei, Z., Leonhardt, N., Torres, M., Dangl, J., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. doi: 10.1093/emboj/cdg277
Li, W., Cong, Q., Kinch, L., and Grishin, N. (2013). Seq2Ref: a web server to facilitate functional interpretation. BMC Bioinformatics 14:30. doi: 10.1186/1471-2105-14-30
Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., et al. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584. doi: 10.1093/nar/gkv279
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulator of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
MacAllister, C., Ohashi-Ito, K., and Bergmann, D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540. doi: 10.1038/nature05491
Males, J., and Griffiths, H. (2017). Stomatal Biology of CAM Plants. Plant Physiol. 174, 550–560. doi: 10.1104/pp.17.00114
McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., et al. (2013). Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600. doi: 10.1093/nar/gkt376
Mhamdi, A., Noctor, G., and Baker, A. (2012). Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys. 525, 181–194. doi: 10.1016/j.abb.2012.04.015
Mhamdi, A., Queval, G., Chaouch, S., Vanderauwera, S., Van Breusegem, F., and Noctor, G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220. doi: 10.1093/jxb/erq282
Mockler, T. C., Michael, T. P., Priest, H. D., Shen, R., Sullivan, C. M., Givan, S. A., et al. (2007). The diurnal project: diurnal and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold. Spring Harb. Symp. Quant. Biol 72, 353–363. doi: 10.1101/sqb.2007.72.006
Moseley, R. C., Mewalal, R., Motta, F., Tuskan, G. A., Haase, S., and Yang, X. (2018). Conservation and diversification of circadian rhythmicity between a model crassulacean acid metabolism plant kalanchoe fedtschenkoi and a Model C3 photosynthesis plant Arabidopsis thaliana. Front. Plant Sci. 9:1757. doi: 10.3389/fpls.2018.01757
Murata, Y., Pei, Z., Mori, I. C., and Schroeder, J. I. (2001). Abscisic acid activation of plasma membrane ca2 channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13, 2513–2523. doi: 10.1105/tpc.13.11.2513
Niu, L., and Liao, W. (2016). Hydrogen peroxide signaling in plant development and abiotic responses: crosstalk with nitric oxide and calcium. Front. Plant Sci. 7:230. doi: 10.3389/fpls.2016.00230
Ohashi-Ito, K., and Bergmann, D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505. doi: 10.1105/tpc.106.046136
Panchy, N., Lehti-Shiu, M., and Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Pearson, W. (2013). An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinformatics 42, 3.1.1–3.1.8. doi: 10.1002/0471250953.bi0301s42
Pei, Z., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Pillitteri, L. J., Sloan, D., GBogenschutz, N., and Torli, K. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505. doi: 10.1038/nature05467
Price, M. N., and Arkin, A. P. (2017). PaperBLAST: text mining papers for information about homologs. mSystems 2:e00039-17. doi: 10.1128/mSystems.00039-17
Queval, G., Issakidis-Bourguet, E., Hoeberichts, F. A., Vandorpe, M., Gakiere, B., Vanacker, H., et al. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657. doi: 10.1111/j.1365-313X.2007.03263.x
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Raissig, M. T., Abrash, E., Bettadapur, A., Vogel, J. P., and Bergmann, D. C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. U.S.A. 113, 8326–8331. doi: 10.1073/pnas.1606728113
Raissig, M. T., Matos, J., Gil, M., Kornfeld, A., Bettadapur, A., Abrash, E., et al. (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355, 1215–1218. doi: 10.1126/science.aal3254
Sewelam, N., Jaspert, N., Van Der Kelen, K., Tognetti, V. B., Schmitz, J., Frerigmann, H., et al. (2014). Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant 7, 1191–1210. doi: 10.1093/mp/ssu070
Stabenau, H., and Winkler, U. (2005). Glycolate metabolism in green algae. Physiol. Plant. 123, 235–245. doi: 10.1111/j.1399-3054.2005.00442.x
Trapnell, C., Robert, A., Goff, L., Pertea, G., Kim, D., Kelley, D., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Van Bel, M., Diels, T., Vancaester, E., Kreft, L., Botzki, A., Van de Peer, Y., et al. (2017). PLAZA 4.0: an integrative resource for functional evolutionary and comparative plant genomices. Nucleic Acids Res. 46, D1190–D1196. doi: 10.1093/nar/gkx1002
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8:161. doi: 10.3389/fpls.2017.00161
Vogel, J. P. (2002). PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell Online 14, 2095–2106. doi: 10.1105/tpc.003509
Woolfenden, H. C., Bourdais, G., Kopischke, M., Miedes, E., Molina, A., Robatzek, S., et al. (2017). A computational approach for inferring the cell wall properties that govern guard cell dynamics. Plant J. 92, 5–18. doi: 10.1111/tpj.13640
Yang, X., Cushman, J. C., Borland, A. M., Edwards, E. J., Wullschleger, S. D., Tuskan, G. A., et al. (2015). A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 207, 491–504. doi: 10.1111/nph.13393
Yang, X., Hu, R., Yin, H., Jenkins, J., Shu, S., Tang, H., et al. (2017). The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 8, 1–15. doi: 10.1038/s41467-017-01491-7
Yang, X., Tuskan, G. A., and Cheng, M. Z. (2006). Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 142, 820–830. doi: 10.1104/pp.106.083642
Zhong, H., and McClung, C. (1996). The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol. Gen. Genet. 251, 196–203.
Keywords: Kalanchoë fedtschenkoi, stomatal movement, crassulacean acid metabolism, drought, rescheduled gene expression, Arabidopsis thaliana, Solanum lycopersicum
Citation: Moseley RC, Tuskan GA and Yang X (2019) Comparative Genomics Analysis Provides New Insight Into Molecular Basis of Stomatal Movement in Kalanchoë fedtschenkoi. Front. Plant Sci. 10:292. doi: 10.3389/fpls.2019.00292
Received: 31 August 2018; Accepted: 22 February 2019;
Published: 13 March 2019.
Edited by:
Dirk Walther, Max-Planck-Institut für Molekulare Pflanzenphysiologie, GermanyReviewed by:
Caspar Christian Cedric Chater, The University of Sheffield, United KingdomHelenice Mercier, University of São Paulo, Brazil
Copyright © 2019 Moseley, Tuskan and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohan Yang, yangx@ornl.gov
 Robert C. Moseley
Robert C. Moseley Gerald A. Tuskan2,3
Gerald A. Tuskan2,3 Xiaohan Yang
Xiaohan Yang