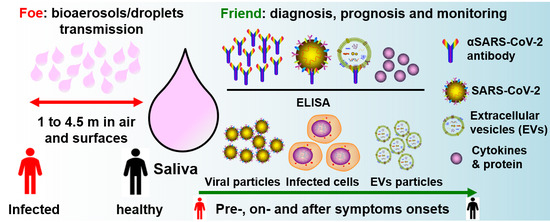
Saliva—Friend and Foe in the COVID-19 Outbreak
Abstract
:1. COVID-19 Pandemic
2. What is SARS-CoV-2?
3. Transmission of COVID-19
4. Current COVID-19 Diagnosis
5. Salivary Droplets and Bioaerosols: A Hidden Foe in COVID-19
6. Salivary Diagnosis and Monitoring: The Friends in Combating COVID-19
6.1. The Versatility of Saliva for COVID-19 Diagnosis
6.2. Salivary Diagnostics for COVID-19
6.3. Salivary Immunity Monitoring for COVID-19
7. Summary: Saliva as Friend and Foe
Funding
Conflicts of Interest
References
- WHO. Available online: http://www.euro.who.int/en/health-topics/health-emergencies/international-health-regulations/news/news/2020/2/2019-ncov-outbreak-is-an-emergency-of-international-concern (accessed on 31 January 2020).
- WHO. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ (accessed on 21 April 2020).
- Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). ArcGIS. Johns Hopkins CSSE. Available online: https://coronavirus.jhu.edu/map.html (accessed on 6 April 2020).
- Max Roser, H.R.A.E.O.-O. Coronavirus Disease (COVID-19)–Statistics and Research. Published online at OurWorldInData.org. Available online: https://ourworldindata.org/coronavirus (accessed on 21 April 2020).
- Hui, D.S.; Azhar, E.E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopkins, C.L.E. Nose and Throat Surgery Body of United Kingdom. Available online: https://www.entuk.org/ (accessed on 28 March 2020).
- China Reveals 1541 Symptom-Free Virus Cases under Pressure. Available online: www.bloomberg.com (accessed on 31 March 2020).
- Nishiura, H.; Kobayashi, T.; Yang, Y.; Hayashi, K.; Miyama, T.; Kinoshita, R.; Linton, N.M.; Jung, S.-M.; Yuan, B.; Suzuki, A.; et al. The Rate of Underascertainment of Novel Coronavirus (2019-nCoV) Infection: Estimation Using Japanese Passengers Data on Evacuation Flights. J. Clin. Med. 2020, 9, 419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveill 2020, 25, 2000180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, A.; Vaquette, C.; Hashimi, S.; Rathnayake, I.; Huygens, F.; Hutmacher, D.W.; Ivanovski, S. Antimicrobial and Immunomodulatory Surface-Functionalized Electrospun Membranes for Bone Regeneration. Adv. Healthc. Mater. 2017, 6, 1345. [Google Scholar] [CrossRef]
- Day, M. Covid-19: Identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ 2020, 368, m1165. [Google Scholar] [CrossRef] [Green Version]
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of viruses from a high proportion of patients with colds. Lancet 1966, 1, 76–77. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.; Osterhaus, A.; Fouchier, R. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, 20. [Google Scholar] [CrossRef] [Green Version]
- National Research Council. Rapid Expert Consultation on the Possibility of Bioaerosol Spread of SARS-CoV-2 for the COVID-19 Pandemic; The National Academies Press: Washington, DC, USA, 2020. [Google Scholar] [CrossRef]
- Morawska, L.; Cao, J. Airborne transmission of SARS-CoV-2: The world should face the reality. Environ. Int. 2020, 139, 105730. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; A Perera, M.R.; Hui, K.P.Y.; Yen, H.-L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef]
- Xia, J.; Tong, J.; Liu, M.; Shen, Y.; Guo, D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020, 92, 589–594. [Google Scholar] [CrossRef] [Green Version]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.-T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic Features and Clinical Course of Patients Infected with SARS-CoV-2 in Singapore. JAMA 2020. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.-I.; Kim, S.-G.; Kim, S.-M.; Kim, E.-H.; Park, S.-J.; Yu, K.-M.; Chang, J.-H.; Lee, S.; Casel, M.A.B.; Um, J.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020. [Google Scholar] [CrossRef]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. SALIVA IS A RELIABLE TOOL TO DETECT SARS-CoV-2. J. Infect. 2020. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Delamater, P.L.; Street, E.; Leslie, T.F.; Yang, Y.T.; Jacobsen, K.H. Complexity of the Basic Reproduction Number (R0). Emerg. Infect. Dis. 2019, 25, 1–4. [Google Scholar] [CrossRef] [Green Version]
- ACR Recommendations for the Use of Chest Radiography and Computed Tomography (CT) for Suspected COVID-19 Infection; American College of Radiology: Reston, VA, USA, 2020.
- Lan, L.; Xu, D.; Ye, G.; Xia, C.; Wang, S.; Li, Y.; Xu, H. Positive RT-PCR Test Results in Patients Recovered from COVID-19. JAMA 2020. [Google Scholar] [CrossRef] [Green Version]
- Tellier, R. Review of aerosol transmission of influenza a virus. Emerg. Infect. Dis. 2006, 12, 1657–1662. [Google Scholar] [CrossRef]
- Wang, J.; Du, G. COVID-19 may transmit through aerosol. Ir. J. Med. Sci. 2020, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, N.-H.W.; Tan, Y.; Taculod, J.; Gorospe, B.; Teope, A.S.; Somani, J.; Tan, A.Y.H. The impact of high-flow nasal cannula (HFNC) on coughing distance: Implications on its use during the novel coronavirus disease outbreak. Can. J. Anaesth. 2020, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Hua, F.; Bian, Z. Coronavirus Disease 2019 (COVID-19): Emerging and Future Challenges for Dental and Oral Medicine. J. Dent. Res. 2020, 22034520914246. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Xu, X.; Li, Y.; Cheng, L.; Zhou, X.; Ren, B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral Sci. 2020, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Sabino-Silva, R.; Jardim, A.C.G.; Siqueira, W.L. Coronavirus COVID-19 impacts to dentistry and potential salivary diagnosis. Clin. Oral Investig. 2020, 24, 1619–1621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dave, M.; Seoudi, N.; Coulthard, P. Urgent dental care for patients during the COVID-19 pandemic. Lancet 2020, 395, 1257. [Google Scholar] [CrossRef]
- Harrel, S.K.; Molinari, J. Aerosols and splatter in dentistry: A brief review of the literature and infection control implications. J. Am. Dent. Assoc. 2004, 135, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.H.L.; Chu, D.K.W.; Shiu, E.Y.C.; Chan, K.-H.; McDevitt, J.J.; Hau, B.J.P.; Yen, H.-L.; Li, Y.; Ip, D.K.M.; Peiris, J.S.M.; et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Pfaffe, T.; Cooper-White, J.J.; Beyerlein, P.; Kostner, K.; Punyadeera, C. Diagnostic potential of saliva: Current state and future applications. Clin. Chem. 2011, 57, 675–687. [Google Scholar] [CrossRef] [Green Version]
- Buduneli, N.; Kinane, D.F. Host-derived diagnostic markers related to soft tissue destruction and bone degradation in periodontitis. J. Clin. Periodontol. 2011, 38 (Suppl. 11), 85–105. [Google Scholar] [CrossRef] [PubMed]
- Kinane, D.F.; P, P.M.; Loos, B.G. Host-response: Understanding the cellular and molecular mechanisms of hostmicrobial interactions: Consensus of the 7th European Workshop on Periodontology. J. Clin. Periodontol. 2011, 38, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.A.; Garlet, G.P.; Fukada, S.Y.; Silva, J.S.; Cunha, F.Q. Chemokines in oral inflammatory diseases: Apical periodontitis and periodontal disease. J. Dent. Res. 2007, 86, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Ebersole, J.; Schuster, J.L.; Stevens, J.; Dawson, D.; Kryscio, R.J.; Lin, Y.; Thomas, M.V.; Miller, C.S. Patterns of salivary analytes provide diagnostic capacity for distinguishing chronic adult periodontitis from health. J. Clin. Immunol. 2013, 33, 271–279. [Google Scholar] [CrossRef]
- Giannobile, W.V.; Al-Shammari, K.F.; Sarment, D.P. Matrix molecules and growth factors as indicators of periodontal disease activity. Periodontology 2003, 31, 125–134. [Google Scholar] [CrossRef]
- Jiang, W.-W.; Masayesva, B.; Zahurak, M.; Carvalho, A.L.; Rosenbaum, E.; Mambo, E.; Zhou, S.; Minhas, K.; Benoit, N.; Westra, W.H.; et al. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin. Cancer Res. 2005, 11, 2486–2491. [Google Scholar] [CrossRef] [Green Version]
- Garbieri, T.F.; Brozoski, D.T.; Dionísio, T.J.; Santos, C.F.; Neves, L. Human DNA extraction from whole saliva that was fresh or stored for 3, 6 or 12 months using five different protocols. J. Appl. Oral Sci. 2017, 25, 147–158. [Google Scholar] [CrossRef] [Green Version]
- Takeshita, T.; Kageyama, S.; Furuta, M.; Tsuboi, H.; Takeuchi, K.; Shibata, Y.; Shimazaki, Y.; Akifusa, S.; Ninomiya, T.; Kiyohara, Y.; et al. Bacterial diversity in saliva and oral health-related conditions: The Hisayama Study. Sci. Rep. 2016, 6, 22164. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wei, Q.; Alvarez, X.; Wang, H.; Du, Y.; Zhu, H.; Jiang, H.; Zhou, J.; Lam, P.; Zhang, L.; et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J. Virol. 2011, 85, 4025–4030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahamtan, A.; Inchley, C.S.; Marzban, M.; Tavakoli-Yaraki, M.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. The role of microRNAs in respiratory viral infection: Friend or foe? Rev. Med. Virol. 2016, 26, 389–407. [Google Scholar] [CrossRef]
- Cristaldi, M.; Mauceri, R.; Di Fede, O.; Giuliana, G.; Campisi, G.; Panzarella, V. Salivary Biomarkers for Oral Squamous Cell Carcinoma Diagnosis and Follow-Up: Current Status and Perspectives. Front. Physiol. 2019, 10, 1476. [Google Scholar] [CrossRef] [PubMed]
- Chiappin, S.; Antonelli, G.; Gatti, R.; De Palo, E.F. Saliva specimen: A new laboratory tool for diagnostic and basic investigation. Clin. Chim. Acta 2007, 383, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Pandit, P.; Cooper-White, J.; Punyadeera, C. High-yield RNA-extraction method for saliva. Clin. Chem. 2013, 59, 1118–1122. [Google Scholar] [CrossRef] [Green Version]
- Han, P.; Ivanovski, S. Effect of Saliva Collection Methods on the Detection of Periodontium-Related Genetic and Epigenetic Biomarkers—A Pilot Study. Int. J. Mol. Sci. 2019, 20, 4729. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef]
- Wang, W.-K.; Chen, S.-Y.; Liu, I.-J.; Chen, Y.-C.; Chen, H.-L.; Yang, C.-F.; Chen, P.-J.; Yeh, S.-H.; Kao, C.-L.; Huang, L.-M.; et al. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg. Infect. Dis. 2004, 10, 1213–1219. [Google Scholar] [CrossRef]
- Researchers Validate Viability of Saliva in COVID-19 Detection. Available online: https://www.mpo-mag.com/contents/view_breaking-news/2020-04-03/researchers-discover-that-saliva-can-detect-covid-19/ (accessed on 4 March 2020).
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Shao, C.; Huang, J.; Gan, J.; Huang, X.; Bucci, E.; Piacentini, M.; Ippolito, G.; Melino, G. COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 2020, 27, 1451–1454. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Ong, E.Z.; Chan, Y.F.Z.; Leong, W.Y.; Lee, N.M.Y.; Kalimuddin, S.; Mohideen, S.M.H.; Chan, K.S.; Tan, A.T.; Bertoletti, A.; Ooi, E.E.; et al. Antonio Bertoletti, and Jenny Guek Hong Low. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe 2020. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Zumla, A.; Locatelli, F.; Ippolito, G.; Kroemer, G. Coronavirus infections: Epidemiological, clinical and immunological features and hypotheses. Cell Stress 2020, 4, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Severance, E.G.; Bossis, I.; Dickerson, F.B.; Stallings, C.R.; Origoni, A.E.; Sullens, A.; Yolken, R.H.; Viscidi, R.P. Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin. Vaccine Immunol. 2008, 15, 1805–1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, B.; Huang, Y.; Huang, L.; Li, B.; Zheng, Z.; Chen, Z.; Chen, J.; Hu, Q.; Wang, H. Effect of mucosal and systemic immunization with virus-like particles of severe acute respiratory syndrome coronavirus in mice. Immunology 2010, 130, 254–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Cassaniti, I.I.; Novazzi, F.; Giardina, F.; Salivaro, F.; Sachs, M.; Perlini, S.; Bruno, R.; Mojoli, F.; Baldanti, F. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.-Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.-X.; et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]



| Sample Size Age (Years) | Sample Source | Diagnosis Technique | Diagnosis Efficiency in Saliva | Reference | |
|---|---|---|---|---|---|
| To et al. | 10 Female, 13 Male Median: 62 (37–75) | Posterior oropharyngeal saliva | RT-qPCR | 87% of patients were viral positive | [30] |
| To et al. | 5 Female, 7 Male Median: 62.5 (35–75) | Saliva from throat | RT-qPCR | 91.7% of patients were positive | [31] |
| Azzi et al. | 8 Female, 17 Male Mean ± standard deviation: 61.5 ± 11.2 | Drooling saliva | RT-qPCR | 100% of patients were viral positive | [33] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, P.; Ivanovski, S. Saliva—Friend and Foe in the COVID-19 Outbreak. Diagnostics 2020, 10, 290. https://doi.org/10.3390/diagnostics10050290
Han P, Ivanovski S. Saliva—Friend and Foe in the COVID-19 Outbreak. Diagnostics. 2020; 10(5):290. https://doi.org/10.3390/diagnostics10050290
Chicago/Turabian StyleHan, Pingping, and Sašo Ivanovski. 2020. "Saliva—Friend and Foe in the COVID-19 Outbreak" Diagnostics 10, no. 5: 290. https://doi.org/10.3390/diagnostics10050290