CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Localization of Target Genes

2.2. Analysis of Dnaic1 Double Point Mutation Phenotype
2.2.1. Generation of Dnaic1 Mutant Mice

2.2.2. Fertility Analysis of Dnaic1 Mutant Mice
2.2.3. Sperm Motility Analysis of Dnaic1 Mutant Mice
2.3. Analysis of Wdr63 sKO Phenotype
2.3.1. Generation of Wdr63 KO Mice

2.3.2. Fertility Analysis of Wdr63 KO Mice
2.3.3. Sperm Motility of Wdr63 KO Mice
2.3.4. Expression Pattern of Wdr78
2.4. Analysis of Ccdc63 sKO Phenotype
2.4.1. Generation of Ccdc63 KO Mice

2.4.2. Fertility Analysis of Ccdc63 KO Mice
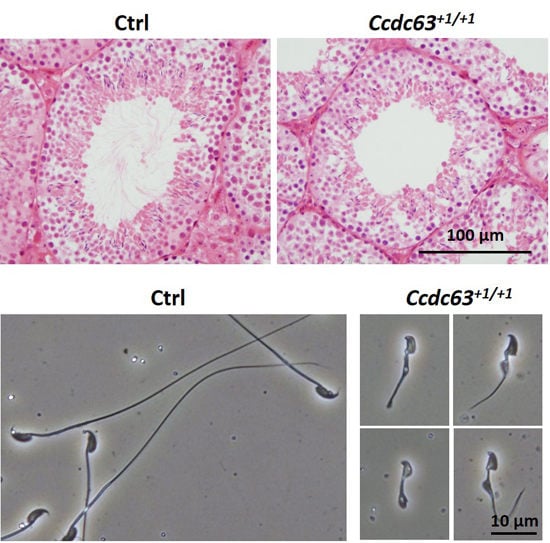
2.4.3. Morphological Analysis of Ccdc63 KO Testes and Spermatozoa

2.5. Discussion
3. Experimental Section
3.1. Animals
3.2. RT-PCR to Determine Gene Expression
3.3. Plasmid and Oligonucleotide Preparation
3.4. Pronuclear Injection
3.5. Genotyping
3.6. Off-Target Analysis of Ccdc63
3.7. Male Fertility Test
3.8. ICSI
3.9. Sperm Motility Analysis
3.10. Visualization of Ccdc63 Null Testes and Spermatozoa
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yanagimachi, R. Mammalian Fertilization; Raven Press: New York, NY, USA, 1994; pp. 189–317. [Google Scholar]
- Inaba, K. Sperm flagella: Comparative and phylogenetic perspectives of protein components. Mol. Hum. Reprod. 2011, 17, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Inaba, K. Molecular architecture of the sperm flagella: Molecules for motility and signaling. Zool. Sci. 2003, 20, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Milisav, I. Dynein and dynein-related genes. Cell Motil. Cytoskelet. 1998, 39, 261–272. [Google Scholar] [CrossRef]
- Pennarun, G.; Escudier, E.; Chapelin, C.; Bridoux, A.M.; Cacheux, V.; Roger, G.; Clément, A.; Goossens, M.; Amselem, S.; Duriez, B. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 dyskinesia. Am. J. Hum. Genet. 1999, 65, 1508–1519. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, R.; Yagi, T. Functional diversity of axonemal dyneins as assessed by in vitro and in vivo motility assays of Chlamydomonas mutants. Zool. Sci. 2014, 31, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Escudier, E.; Duquesnoy, P.; Papon, J.F.; Amselem, S. Ciliary defects and genetics of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2009, 10, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Harricane, M.C.; Lafitte, J.J.; Godard, P.; Zaegel, M.; Tack, V.; Lalau, G.; Bouvagnet, P. Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome). Am. J. Hum. Genet. 2001, 68, 1030–1035. [Google Scholar] [CrossRef] [PubMed]
- Onoufriadis, A.; Paff, T.; Antony, D.; Shoemark, A.; Micha, D.; Kuyt, B.; Schmidts, M.; Petridi, S.; Dankert-Roelse, J.E.; Haarman, E.G.; et al. Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 92, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Zariwala, M.; Noone, P.G.; Sannuti, A.; Minnix, S.; Zhou, Z.; Leigh, M.W.; Hazucha, M.; Carson, J.L.; Knowles, M.R. Germline mutations in an intermediate chain dynein cause primary ciliary dyskinesia. Am. J. Respir. Cell Mol. Biol. 2001, 25, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Zariwala, M.A.; Leigh, M.W.; Ceppa, F.; Kennedy, M.P.; Noone, P.G.; Carson, J.L.; Hazucha, M.J.; Lori, A.; Horvath, J.; Olbrich, H.; et al. Mutations of DNAI1 in primary ciliary dyskinesia: Evidence of founder effect in a common mutation. Am. J. Respir. Crit. Care Med. 2006, 174, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Munro, N.C.; Currie, D.C.; Lindsay, K.S.; Ryder, T.A.; Rutman, A.; Dewar, A.; Greenstone, M.A.; Hendry, W.F.; Cole, P.J. Fertility in men with primary ciliary dyskinesia presenting with respiratory infection. Thorax 1994, 49, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Mashiko, D.; Fujihara, Y.; Satouh, Y.; Miyata, H.; Isotani, A.; Ikawa, M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Mashiko, D.; Young, S.A.; Muto, M.; Kato, H.; Nozawa, K.; Ogawa, M.; Noda, T.; Kim, Y.J.; Satouh, Y.; Fujihara, Y.; et al. Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes. Dev. Growth Differ. 2014, 56, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Damerla, R.R.; Chatterjee, B.; Li, Y.; Francis, R.J.; Fatakia, S.N.; Lo, C.W. Ion torrent sequencing for conducting genome-wide scans for mutation mapping analysis. Mamm. Genome 2014, 25, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.J.; Christopher, A.; Devine, W.A.; Ostrowski, L.; Lo, C. Congenital heart disease and the specification of left-right asymmetry. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H2102–H2111. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, L.E.; Yin, W.; Rogers, T.D.; Busalacchi, K.B.; Chua, M.; O’Neal, W.K.; Grubb, B.R. Conditional deletion of Dnaic1 in a murine model of primary ciliary dyskinesia causes chronic rhinosinusitis. Am. J. Respir. Cell Mol. Biol. 2010, 43, 55–63. [Google Scholar] [CrossRef]
- Baker, M.A.; Hetherington, L.; Weinberg, A.; Naumovski, N.; Velkov, T.; Pelzing, M.; Dolman, S.; Condina, M.R.; Aitken, R.J. Analysis of phosphopeptide changes as spermatozoa acquire functional competence in the epididymis demonstrates changes in the post-translational modification of Izumo1. J. Proteome Res. 2012, 11, 5252–5264. [Google Scholar] [PubMed]
- Baker, M.A.; Smith, N.D.; Hetherington, L.; Taubman, K.; Graham, M.E.; Robinson, P.J.; Aitken, R.J. Label-free quantitation of phosphopeptide changes during rat sperm capacitation. J. Proteome Res. 2010, 9, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Platt, M.D.; Salicioni, A.M.; Hunt, D.F.; Visconti, P.E. Use of differential isotopic labeling and mass spectrometry to analyze capacitation-associated changes in the phosphorylation status of mouse sperm proteins. J. Proteome Res. 2009, 8, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Sale, W.S. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 1998, 9, 3335–3349. [Google Scholar] [PubMed]
- Kamiya, R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 2002, 219, 115–155. [Google Scholar] [PubMed]
- Hozumi, A.; Padma, P.; Toda, T.; Ide, H.; Inaba, K. Molecular characterization of axonemal proteins and signaling molecules responsible for chemoattractant-induced sperm activation in Ciona intestinalis. Cell Motil. Cytoskelet. 2008, 65, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Murata, M.; Inaba, K.; Morisawa, M. A chemoattractant for ascidian spermatozoa is a sulfated steroid. Proc. Natl. Acad. Sci. USA 2002, 99, 14831–14836. [Google Scholar] [PubMed]
- Owa, M.; Furuta, A.; Usukura, J.; Arisaka, F.; King, S.M.; Witman, G.B.; Kamiya, R.; Wakabayashi, K. Cooperative binding of the outer arm-docking complex underlies the regular arrangement of outer arm dynein in the axoneme. Proc. Natl. Acad. Sci. USA 2014, 111, 9461–9466. [Google Scholar] [CrossRef] [PubMed]
- Elliott, B.; Richardson, C.; Winderbaum, J.; Nickoloff, J.A.; Jasin, M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1998, 18, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Miyado, M.; Igarashi, M.; Tamano, M.; Kubo, A.; Yamashita, S.; Asahara, H.; Fukami, M.; Takada, S. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 2014, 4, 5396. [Google Scholar] [CrossRef] [PubMed]
- Perrone, C.A.; Yang, P.; O’Toole, E.; Sale, W.S.; Porter, M.E. The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol. Biol. Cell 1998, 9, 3351–3365. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.W.; Craige, B.; Kroeger, T.V.; Finn, R.; Wyatt, T.A.; Sisson, J.H.; Pavlik, J.A.; Strittmatter, L.; Hendricks, G.M.; Witman, G.B.; et al. CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Mol. Biol. Cell 2015, 26, 3140–3149. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Watanabe, M.; Okada, Y.; Sawa, H.; Takai, H.; Nakanishi, M.; Kawase, Y.; Suzuki, H.; Nagashima, K.; Ikeda, K.; et al. Hydrocephalus, situs inversus, chronic sinusitis, and male infertility in DNA polymerase lambda-deficient mice: Possible implication for the pathogenesis of immotile cilia syndrome. Mol. Cell. Biol. 2002, 22, 2769–2776. [Google Scholar] [CrossRef] [PubMed]
- Zariwala, M.; O’Neal, W.K.; Noone, P.G.; Leigh, M.W.; Knowles, M.R.; Ostrowski, L.E. Investigation of the possible role of a novel gene, DPCD, in primary ciliary dyskinesia. Am. J. Respir. Cell Mol. Biol. 2004, 30, 428–434. [Google Scholar] [PubMed]
- Zhou, J.; Yang, F.; Leu, N.A.; Wang, P.J. MNS1 is essential for spermiogenesis and motile ciliary functions in mice. PLoS Genet. 2012, 8, e1002516. [Google Scholar] [CrossRef] [PubMed]
- Ferlin, A.; Rocca, M.S.; Vinanzi, C.; Ghezzi, M.; di Nisio, A.; Foresta, C. Mutational screening of NR5A1 gene encoding steroidogenic factor 1 in cryptorchidism and male factor infertility and functional analysis of seven undescribed mutations. Fertil. Steril. 2015, 104, 163–169. e161. [Google Scholar] [PubMed]
- Sato, Y.; Tajima, A.; Tsunematsu, K.; Nozawa, S.; Yoshiike, M.; Koh, E.; Kanaya, J.; Namiki, M.; Matsumiya, K.; Tsujimura, A.; et al. An association study of four candidate loci for human male fertility traits with male infertility. Hum. Reprod. 2015, 30, 1510–1514. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, Y.; Nishimura, H.; Tsujimura, A.; Matsuoka, Y.; Matsumiya, K.; Okuyama, A.; Nishimune, Y.; Tanaka, H. Single-nucleotide polymorphisms and mutation analyses of the TNP1 and TNP2 genes of fertile and infertile human male populations. J. Androl. 2005, 26, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Nishimune, Y.; Tanaka, H. Infertility caused by polymorphisms or mutations in spermatogenesis-specific genes. J. Androl. 2006, 27, 326–334. [Google Scholar] [PubMed]
- Fujihara, Y.; Murakami, M.; Inoue, N.; Satouh, Y.; Kaseda, K.; Ikawa, M.; Okabe, M. Sperm equatorial segment protein 1, SPESP1, is required for fully fertile sperm in mouse. J. Cell Sci. 2010, 123, 1531–1536. [Google Scholar] [PubMed]
- Bowtie: An ultrafast memory-efficient short read aligner; Johns Hopkins University. Available online: http://bowtie-bio.sourceforge.net/index.shtml (accessed on 16 July 2013).
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Shivalila, C.; Cheng, A.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Wigglesworth, K.; Eppig, J.J.; Schultz, R.M. Preimplantation development of mouse embryos in KSOM: Augmentation by amino acids and analysis of gene expression. Mol. Reprod. Dev. 1995, 41, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, Y.; Kaseda, K.; Inoue, N.; Ikawa, M.; Okabe, M. Production of mouse pups from germline transmission-failed knockout chimeras. Transgenic Res. 2013, 22, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Yanagimachi, R. Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development 1995, 121, 2397–2405. [Google Scholar] [PubMed]
- Toyoda, Y.; Yokoyama, M.; Hoshi, T. Studies on the fertilization of mouse eggs in vitro. Jpn. J. Anim. Reprod. 1971, 16, 147–151. [Google Scholar] [CrossRef]
- Goodson, S.G.; Zhang, Z.; Tsuruta, J.K.; Wang, W.; O’Brien, D.A. Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol. Reprod. 2011, 84, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Inoue, N.; Satouh, Y.; Ikawa, M.; Okabe, M.; Yanagimachi, R. Acrosome-reacted mouse spermatozoa recovered from the perivitelline space can fertilize other eggs. Proc. Natl. Acad. Sci. USA 2011, 108, 20008–20011. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Young, S.A.M.; Miyata, H.; Satouh, Y.; Kato, H.; Nozawa, K.; Isotani, A.; Aitken, R.J.; Baker, M.A.; Ikawa, M. CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis. Int. J. Mol. Sci. 2015, 16, 24732-24750. https://doi.org/10.3390/ijms161024732
Young SAM, Miyata H, Satouh Y, Kato H, Nozawa K, Isotani A, Aitken RJ, Baker MA, Ikawa M. CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis. International Journal of Molecular Sciences. 2015; 16(10):24732-24750. https://doi.org/10.3390/ijms161024732
Chicago/Turabian StyleYoung, Samantha A. M., Haruhiko Miyata, Yuhkoh Satouh, Hirotaka Kato, Kaori Nozawa, Ayako Isotani, R. John Aitken, Mark A. Baker, and Masahito Ikawa. 2015. "CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis" International Journal of Molecular Sciences 16, no. 10: 24732-24750. https://doi.org/10.3390/ijms161024732