Two Novel Relative Double-Stranded RNA Mycoviruses Infecting Fusarium poae Strain SX63
Abstract
:1. Introduction
2. Results and Discussion
2.1. Detection and Complete Genome Sequencing of dsRNA in Fusarium poae Strain SX630
2.2. Both FpV2 and FpV3 Have a Double-Stranded RNA Genome
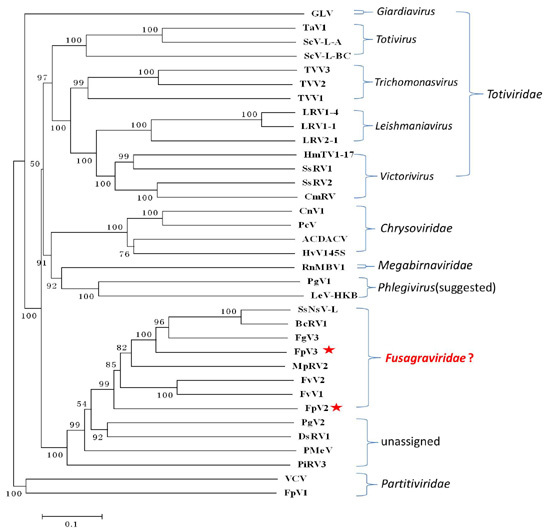
2.3. Phylogenetic Analysis Based on the RdRp and P1 Sequences
2.4. Identification of “Phytoreo_S7 Domain” in FpV2, FpV3, and Related Unassigned dsRNA Viruses
2.5. Potential Programmed –1 Ribosomal Frameshifting (–1 PRF) in FpV2, FpV3, and Related Unassigned dsRNA Viruses
2.6. A Proposition to Create a New Family Designated Fusagraviridae
3. Materials and Methods
3.1. Fungal Strain and Culture Conditions
3.2. dsRNA Extraction and Purification
3.3. cDNA Synthesis, Molecular Cloning, and Sequencing
3.4. Nucleotide Sequence Analysis
3.5. Phylogenetic Analysis
3.6. GenBank Accession Number
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, B.; Fu, Y.; Jiang, D.; Ghabrial, S.A.; Li, G.; Peng, Y.; Xie, J.; Cheng, J.; Huang, J.; et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Chiba, S.; Toyoda, K.; Suzuki, N. Evidence for negative-strand RNA virus infection in fungi. Virology 2013, 435, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xie, J.; Cheng, J.; Fu, Y.; Li, G.; Yi, X.; Jiang, D. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc. Natl. Acad. Sci. USA 2014, 111, 12205–12210. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier Academic Press: London, UK, 2012; Volume 9, pp. 23–1208. [Google Scholar]
- Yu, L.; Sang, W.; Wu, M.D.; Zhang, J.; Yang, L.; Zhou, Y.J.; Chen, W.D.; Li, G.Q. Novel hypovirulence-associated RNA mycovirus in the plant-pathogenic fungus Botrytis cinerea: Molecular and biological characterization. Appl. Environ. Microbiol. 2015, 81, 2299–2310. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Kwon, S.J.; Lee, K.M.; Son, M.; Kim, K.H. Complete nucleotide sequence of double-stranded RNA viruses from Fusarium graminearum strain DK3. Arch. Virol. 2009, 154, 1855–1858. [Google Scholar] [CrossRef] [PubMed]
- Marvelli, R.A.; Hobbs, H.A.; Li, S.; McCoppin, N.K.; Domier, L.L.; Hartman, G.L.; Eastburn, D.M. Identification of novel double-stranded RNA mycoviruses of Fusarium virguliforme and evidence of their effects on virulence. Arch. Virol. 2014, 159, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fu, Y.; Xie, J.; Cheng, J.; Ghabrial, S.A.; Li, G.; Peng, Y.; Yi, X.; Jiang, D. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol. Biol. 2012, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.N.; Beever, R.E.; Boine, B.; Arthur, K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009, 10, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Salaipeth, L.; Lin, Y.H.; Sasaki, A.; Kanematsu, S.; Suzuki, N. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: Molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 2009, 83, 12801–12812. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Hypovirulence: Mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Anagnostakis, S.L. Biological control of chestnut blight. Science 1982, 215, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.; Jenkinson, P.; McLeod, L. Fusarium ear blight (scab) in small grain cereals—A review. Plant Pathol. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Rocha, O.; Ansari, K.; Doohan, F.M. Effects of trichothecene mycotoxins on eukaryotic cells: A review. Food Addit. Contam. 2005, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Fekete, C.; Giczey, G.; Papp, I.; Szabo, L.; Hornok, L. High-frequency occurrence of virus-like particles with double-stranded RNA genome in Fusarium poae. FEMS Microbiol. Lett. 1995, 131, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Compel, P.; Papp, I.; Bibo, M.; Fekete, C.; Hornok, L. Genetic interrelationships and genome organization of double-stranded RNA elements of Fusarium poae. Virus Genes 1999, 18, 49–56. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Kistler, H.C.; Tacke, B.K.; Casper, H.H. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 2000, 97, 7905–7910. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, H.; Chen, X.; Qiu, D.; Guo, L. Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 2015, 481, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Urbez-Torres, J.R.; Cordero, F.; Rowhani, A. Deep sequencing evidence from single grapevine plants reveals a virome dominated by mycoviruses. Arch. Virol. 2011, 156, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Kozlakidis, Z.; Hacker, C.V.; Bradley, D.; Jamal, A.; Phoon, X.; Webber, J.; Brasier, C.M.; Buck, K.W.; Coutts, R.H. Molecular characterisation of two novel double-stranded RNA elements from Phlebiopsis gigantea. Virus Genes 2009, 39, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Abreu, E.F.; Daltro, C.B.; Nogueira, E.O.; Andrade, E.C.; Aragão, F.J. Sequence and genome organization of papaya meleira virus infecting papaya in Brazil. Arch. Virol. 2015, 160, 3143–3147. [Google Scholar] [CrossRef] [PubMed]
- De Wet, J.; Bihon, W.; Preisig, O.; Wingfield, B.D.; Wingfield, M.J. Characterization of a novel dsRNA element in the pine endophytic fungus Diplodia scrobiculata. Arch. Virol. 2011, 156, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Krychiw, J.F.; Myers, K.; Fry, W.E.; Hillman, B.I. A new virus from the plant pathogenic oomycete Phytophthora infestans with an 8 kb dsRNA genome: The sixth member of a proposed new virus genus. Virology 2013, 435, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Bruenn, J.A. A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res. 1993, 21, 5667–5669. [Google Scholar] [CrossRef] [PubMed]
- Petrzik, K.; Sarkisova, T.; Starý, J.; Koloniuk, I.; Hrabáková, L.; Kubešová, O. Molecular characterization of a new monopartite dsRNA mycovirus from mycorrhizal Thelephora terrestris (Ehrh.) and its detection in soil oribatid mites (Acari: Oribatida). Virology 2015, 489, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.L.; Atkins, J.F.; Gesteland, R.F. Programmed ribosomal frameshifting: Much ado about knotting. Proc. Natl. Acad. Sci. USA 1999, 96, 14177–14179. [Google Scholar] [CrossRef] [PubMed]
- Caliskan, N.; Peske, F.; Rodnina, M.V. Changed in translation: mRNA recoding by –1 programmed ribosomal frameshifting. Trends Biochem. Sci. 2015, 40, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W.; Miller, W.A. Translational control in positive strand RNA plant viruses. Virology 2006, 44, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Brierley, I.; Gilbert, R.J.C.; Pennell, S. Recoding: Expansion of Decoding Rules Enriches Gene Expression; Atkins, J.F., Gesteland, R.F., Eds.; Springer: New York, NY, USA, 2010; pp. 149–174. [Google Scholar]
- Moon, S.; Byun, Y.; Kim, H.J.; Jeong, S.; Han, K. Predicting genes expressed via –1 and +1 frameshifts. Nucleic Acids Res. 2004, 32, 4884–4892. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.D.; Icho, T.; Wickner, R.B. A –1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. USA 1991, 88, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.; Nameth, S.; Jordan, R.L. Analysis of double-stranded RNA for plant virus diagnosis. Plant Dis. 1990, 74, 255–258. [Google Scholar]
- Wang, S.; Kondo, H.; Liu, L.; Guo, L.; Qiu, D. A novel virus in the family Hypoviridae from the plant pathogenic fungus Fusarium graminearum. Virus Res. 2013, 174, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Xiao, X.; Fu, Y.; Liu, H.; Cheng, J.; Ghabrial, S.A.; Li, G.; Jiang, D. A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 2011, 418, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wei, D.; Jiang, D.; Fu, Y.; Li, G.; Ghabrial, S.; Peng, Y. Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J. Gen. Virol. 2006, 87, 241–249. [Google Scholar] [CrossRef] [PubMed]
- NCBI ORF Finder Tool. Available online: http://www.ncbi.nlm.nih.gov/projects/gorf/ (accessed on 20 January 2016).
- NCBI CDD Search. Available online: http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 20 January 2016).
- NCBI Blast Program. Available online: http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi (accessed on 20 January 2016).
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinfrom. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]





| Virus | Full Sequence | 5′-UTR | 3′-UTR | ORF1 (P1) | ORF2 (RdRP) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | nt % | Length (bp) | nt % | Length (bp) | nt % | nt % | aa % | nt % | aa % | |
| FpV2 | 9518 | 42.97 | 1029 | 36.12 | 48 | 44.07 | 39.91 | 24.27 | 44.05 | 29.44 |
| FpV3 | 9419 | 1190 | 55 | |||||||
| Search Target And Virus Name (GenBank Accession No.) a | Size (aa) | % Identity | Overlap (Positions) | Bit Score | E-Value |
|---|---|---|---|---|---|
| FpV2 (FpV3) | FpV2 (FpV3) | FpV2 (FpV3) | FpV2 (FpV3) | ||
| P1 search | |||||
| MpRV2 (ALD89096) | 1285 | 34 (38) | 322/960 (340/885) | 482 (558) | 1 × 10−142 (9 × 10–172) |
| SsNsV-L (YP_006331064) | 1305 | 33 (49) | 353/1057 (619/1276) | 424 (1125) | 1 × 10–121 (0.0) |
| FgV3 (YP_003288788) | 1369 | 32 (44) | 328/1037 (538/1217) | 423 (944) | 8 × 10−121 (0.0) |
| GaTV2 (ADO60932) | 1313 | 33 (47) | 353/1076 (635/1351) | 397 (1123) | 2 × 10−112 (0.0) |
| BcRV1 (YP_009115497) | 1314 | 32 (47) | 347/1075 (644/1363) | 395 (1118) | 1 × 10−111 (0.0) |
| FvV2 (AEZ54145) | 1347 | 27 (28) | 295/1083 (257/910) | 337 (329) | 6 × 10−92 (1 × 10−89) |
| FvV1 (AEZ54147) | 1311 | 26 (29) | 281/1066 (286/983) | 335 (357) | 1 × 10−91 (5 × 10−99) |
| PgV2 (CAJ34334) b | >1696 | 22 (28) | 195/886 (117/424) | 123 (124) | 9 × 10−25 (4 × 10−25) |
| PmeV (YP_009179229) | 1596 | 25 (ND) | 93/365 (ND) | 91.7 (ND) | 5 × 10−15 (ND) |
| RdRp search | |||||
| MpRV2 (ALD89097) | 1082 | 36 (37) | 386/1076 (410/1116) | 625 (664) | 0.0 (0.0) |
| FgV3 (YP_003288789) | 1311 | 32 (44) | 421/1324 (605/1366) | 568 (1041) | 1 × 10−174 (0.0) |
| FvV1 (AEZ54148) | 1289 | 30 (31) | 401/1328 (408/1305) | 556 (561) | 2 × 10−170 (3 × 10−172) |
| BcRV1 (YP_009115498) | 1338 | 31 (43) | 427/1356 (594/1372) | 556 (993) | 5 × 10−170 (0.0) |
| SsNsV-L (CEZ26308) | 1338 | 31 (44) | 421/1338 (601/1370) | 556 (1056) | 7 × 10−170 (0.0) |
| FvV2 (AEZ54146) | 1310 | 30 (31) | 406/1346 (662/1398) | 535 (558) | 1 × 10−162 (6 × 10−171) |
| DsRV1 (YP_003359178) | 1110 | 28 (32) | 305/1094 (322/1005) | 365 (436) | 8 × 10−103 (5 × 10−128) |
| PgV2 (CAJ34335) | 1153 | 36 (38) | 226/632 (234/623) | 331 (380) | 5 × 10−91 (2 × 10−107) |
| PiRV3 (AEX87902) | 1011 | 29 (31) | 190/645 (188/614) | 243 (241) | 1 × 10−62 (8 × 10−62) |
| PmeV (YP_009179230) | 1156 | 31 (33) | 166/535 (197/604) | 241 (280) | 2 × 10−61 (3 × 10−74) |
| GaTV2 (ADO60933) b | >613 | 31(44) | 196/632 (274/626) | 230 (456) | 3 × 10−60 (6 × 10−141) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhang, J.; Zhang, H.; Qiu, D.; Guo, L. Two Novel Relative Double-Stranded RNA Mycoviruses Infecting Fusarium poae Strain SX63. Int. J. Mol. Sci. 2016, 17, 641. https://doi.org/10.3390/ijms17050641
Wang L, Zhang J, Zhang H, Qiu D, Guo L. Two Novel Relative Double-Stranded RNA Mycoviruses Infecting Fusarium poae Strain SX63. International Journal of Molecular Sciences. 2016; 17(5):641. https://doi.org/10.3390/ijms17050641
Chicago/Turabian StyleWang, Luan, Jingze Zhang, Hailong Zhang, Dewen Qiu, and Lihua Guo. 2016. "Two Novel Relative Double-Stranded RNA Mycoviruses Infecting Fusarium poae Strain SX63" International Journal of Molecular Sciences 17, no. 5: 641. https://doi.org/10.3390/ijms17050641