Genetic Diversity of Ethiopian Durum Wheat (Triticum durum Desf.) Landrace Collections as Reveled by SSR Markers
Received: 08-Nov-2018 / Accepted Date: 28-Jan-2019 / Published Date: 05-Feb-2019 DOI: 10.4172/2329-8863.1000413
Abstract
The genetic diversity of 141 Ethiopian durum wheat (Triticum durum Desf.) landrace collections and 19 improved varieties was analyzed using primers of 12 SSR markers in 2015. The experiment was conducted at the National Agricultural Biotechnology Laboratory, Holetta, Ethiopia. The difference between the longest and shortest amplified fragment size ranged from 100 to 350 bp. The highest variation in fragment size was observed for primer CFD 257 (250-350 bp) and the lowest was for primer CFA2278 (100-180 bp). All the 12 SSR primer pairs were polymorphic and generated a total of 74 alleles. The number of alleles per locus ranged from 4 for primers WMS 375, WMS 493, and CFD257 to 9 for primers WMS53, with a mean of 6 alleles per locus. Some loci were more monomorphic (WMS493, WMS516, and WMS532) with less gene flow than other loci such as WMS269 and WMS234. Based on the analysis of molecular variance (AMOVA), lower level (19%) of variation was observed among populations and higher level (81%) within population. The dendogram of cluster analysis based on neighbor-joining algorithm categorized the 160 durum wheat genotypes into three major clusters. Cluster I consisted 47, Cluster II 64, and Cluster III 49 genotypes. Based on the magnitude of the genetic distance (GD), more differentiations were observed between accessions in populations originated from different geographical regions in Ethiopia.
Keywords: Durum wheat; Genetic diversity; Landrace; SSR marker
Introduction
Durum wheat (Triticum durum Desf.) is a member of the Gramineae family which belongs to the Triticeae tribe. It is an allotetraploid (two genomes: AABB) with a total of 28 chromosomes (2n=4x=28). Triticum durum is believed to be originated thousands of years ago from a hybridization between the wild diploid T. monococcum L. subsp. Boeoticum (Boiss) (A genome donor) (Synonym: Triticumurartu: AA) and the donor of the B genome which, based on morphological, geographical and cytological evidence, has been recognized as T. speltoides (Tausch) Gren or its closely related species [1-3].
Genetic divergence arises either as a result of geographic separation or genetic barriers to cross ability [4]. Knowledge of the extent and pattern of genetic diversity within and between populations (accessions) is very important for the identification of useful materials for plant breeding purposes and to better understand the crop to design appropriate collection and conservation strategies. It is believed that crosses between genetically diverse parents are likely to produce higher hetrosis, desirable genetic recombination and segregation in progenies [4,5].
The inception of wheat breeding in Ethiopia dated back to the early 1960’s and, as the result of the efforts made hitherto, a number of improved varieties have been developed and released to farmers. Ethiopia has also benefited from the country-wide scaling up of improved varieties of a limited number of cereals dominantly maize (Zea mays ) and bread wheat (Triticum aestivum L.) [6]. Modern varieties of bread wheat (Triticum astivum L.) are believed to have substituted durum wheat varieties and it is generally believed that the genetic diversity in durum wheat has been increasingly dwindling [7]. Narrow genetic diversity is a major problem for breeding varieties with better adaptation to different agro-ecologies, resistant/tolerant to biotic stresses like diseases, and abiotic stresses like drought.
Despite the importance of knowledge of genetic diversity in Ethiopian durum wheat landraces for future breeding, the available information is not exhaustive and representative. First, most of the studies on Ethiopian wheat landraces were conducted elsewhere out of their natural habitat [8-10]. In some instances, the samples did not include major production regions like the northern and the eastern parts of the country but just concentrated on the central highlands [10]. Second, in the majority of the past studies, durum wheat was evaluated together with hexaploid wheat and there was no desegregated result [10]. Third, even though some of the studies were inclusive in terms of area coverage and markers used, the studies were conducted long ago and the available information is not contemporary. Therefore, this study was aimed at assessing the magnitude and pattern of genetic diversity in Ethiopian durum wheat landraces collected from the three major durum wheat producing regions (Amhara, Oromia and Tigray) of the country.
Materials and Methods
Plant materials and DNA extraction
A total of 141 durum wheat germplasm accessions and 19 released varieties were used for this experiment (Table 1). Sound seeds of each genotype were planted in the greenhouse of the National Agricultural Biotechnology Laboratory, Holetta, Ethiopia, in 2015. Three weeks after planting, approximately equal amount of bulk leave samples were collected from 3-5 plants of each entry and placed in 50 ml autoclaved and labeled falcon tubes with enough amount of yellow silica gel as a desiccant. After the leaves were totally dried, approximately 50 mg of dry leaves were ground using a mixer mill (Retsch RM 2000) and genomic DNA was extracted using the cetyltriethyl ammonium bromide (CTAB) method [11]. The quality and quantity of the DNA samples were tested using gel pictures and then confirmed with the use of spectro-nano-gram optimizer (ND-8000). Samples with high band intensity and lesser smear were maintained for polymerase chain reaction (PCR) but samples with low quality were suspended for re-extraction.
| Region | Zone | No. of Genotypes | Name of genotypes |
|---|---|---|---|
| Amhara | South Wello | 10 (01-10) | Acc.No: 231623, 231597, 231600, 222855, 226094, 8185, 8186, 214590, 214550, 213149 |
| Amhara | South Gonder | 11 (11-21) | Acc.No: 206573, 222621, 222641, 216614, 222655, 222608, 222613, 7412, 216474, 226951, 222616 |
| Amhara | West Gojam | 6 (22-27) | Acc.No: 203750, 203922, 203893, 5487, 208212, 203757 |
| Amhara | East Gojam | 11 (28-38) | Acc.No: 208189, 208195, 210821,226833,226844, 231617, 231618, 214515, 8333, 8328, 214517 |
| Amhara | North Gonder | 10 (39-48) | Acc.No: 222515, 203840, 5217, 204340, 6856, 216492, 216545, 226207, 226208, 216440 |
| Oromia | North Shewa | 11 (49-59) | Acc.No: 208265, 208278, 208286, 208310, 208312, 208317, 208491, 226375, 5679, 5739, 226892 |
| Oromia | Arsi | 11 (60-70) | Acc.No: 222421, 222422, 222428, 226868, 226356, 7073, 214498, 7022, 226273, 5927 |
| Oromia | Bale | 07 (71-77) | Acc.No: 231467, 222324, 222338, 204349, 204370, 227060, 204357 |
| Oromia | Harerge | 14 (78-91) | Acc.No: 214503, 203695, 203886, 226180, 226183, 222708, 231471, 203690, 231603, 5730, 203854, 226179, 231606, 231613 |
| Oromia | East Shewa | 11 (92-102) | Acc.No: 210808, 5429, 216651, 5314, 203748, 5300, 5248, 5180, 5736, 214313, 226959 |
| Oromia | West shewa | 11 (103-113) | Acc.No: 231528, 222457. 222461, 231557, 231526, 214328, 5454, 5144, 6101, 7206, 227020 |
| Tigray | Southern | 10 (114-123) | Acc.No: 214343, 7956, 207854, 206551, 206554, 223257, 226199, 206558, 238113, 226245 |
| Tigray | Central | 10 (124-133) | Acc.No: 238114, 238118, 238121, 238122, 238123, 238124, 238125, 238126, 238136, 238137 |
| Tigray | Eastern | 08 (134-141) | Acc.No: 238127, 238128, 238129, 238130, 238131, 238133, 238134, 238135 |
| Improved Varieties | 19 (142-160) | Ginchi, Yerer, Worer, Mangudo, Arendato, Assasa, Denbi,Tob, LD-357, Hitosa, Mukye, Killinto, Quamy, Gerardo, Foka, Cocorit, Boohie, Bichena, Meteyaya | |
Table 1: Details of the Durum wheat germplasm accessions used for the study.
Primer selection and optimization
About 53 simple sequence repeat (SSR) primers previously reported to be polymorphic with other sets of durum wheat genetic materials [12-14] were pre-screened for polymorphism in this specific population and then 12 primers which showed high polymorphism were selected for use in PCR (Table 2).
| Microsatellites | Primer sequences | |
|---|---|---|
| Forward | Reverse | |
| CFA2278 2BS | AAGTCGGCCATCTTCTTCCT | GCCTCTGCAAGTCTTTACCG |
| Wms493 3BS | GGAACATCATTTCTGGACTTTG | TTCCCATAACTAAAACCGCG |
| Wmc516 4AS | GACTCGCAACTAGGGGT | GGGCCACGAATAAACAG |
| Wms5 3AL | GCCAGCTACCTCGATACAACTC | AGAAAGGGCCAGGCTAGTAGT |
| Wmc532 3AS | GATACATCAAGATCGTGCCAAA | GGGAGAAATCATTAACGAAGGG |
| Wms120 2BL | GATCCACCTTCCTCTCTCTC | GATTATACTGGTGCCGAAAC |
| WMC516 | GGGCCACGAATAAACAG | GACTCGCAACTAGGGGT |
| WMS269 | TGCATATAAACAGTCACACACCC | TTTGAGCTCCAAAGTGAGTTAGC |
| CFD257 | TCTCAACTTGCAACTGCCAC | CCCTCCATGGATTCTTGCTA |
| BARC153 | CGCGCCTTGCTTTATTAGTATTAGTATT | GCGGCATGCACATATAATTCTCATTGACT |
| WMS234 5BL | GAGTCCTGATGTGAAGCTGTTG | CTCATTGGGGTGTGTACGTG |
| WMS375 4BL | ATTGGCGACTCTAGCATATACG | GGGATGTCTGTTCCATCTTAGC |
| Xgwm1361AS | GACAGCACCTTGCCCTTTG | CATCGGCAACATGCTCATC |
Table 2: Details (forward and reverse sequences) of the polymorphic microsatellite markers used in this study.
Procedures of polymerase chain reaction (PCR)
PCR reaction was performed with a thermal cycler (GeneAmp@PCR System 9700) in a total volume of 25 μl containing 6.25 μl green promega master mix containing all components of PCR except genomic DNA and primers 3.75 μl, 1.25 μl forward and 1.25 μl reverse primer and 12.5 nuclease free water. The PCR was programmed at an initial denaturation step of 2 minutes at 94°C followed by 30 cycles of 30 seconds denaturation at 94°C, annealing at 51°C to 60°C (depending on the requirements of the primers) for 30 seconds, initial extension at 72°C for 1minute and final extension at 72°C for 10 minutes. At the end of the reaction, the PCR products were stored at -200°C and were later subjected to agarose gel ectrophoresis to ensure amplification.
Polyacrylamide gel electrophoresis (PAGE)
Electrophoresis was carried out on a vertical electrophoresis set up (CS-500 V) at a constant voltage of 300 V for 1:30 h using a standard DNA ladder (HyperLadder TM 500-2000 bp) with known reference band to estimate the molecular weight of each amplified product. Amplification was visualized under 3 UV-trans illuminator. Primers with unclear and missing bands were sorted out and repeated.
Data scoring
Clearly resolved and unambiguous bands were scored visually for their presence or absence for each primer and sample [15]. Non-polymorphic, missing, faint and distorted gels were overlooked at scoring and only records of twelve markers with clear polymorphic bands were considered for statistical analysis. Each fragment was scored independently as ‘1’ for presence and ‘0’ for absence. Bands with the same molecular weight were treated as identical fragments. The total numbers of bands, number of polymorphic bands in a set of accessions, and genetic diversity of all individuals and across each primer were calculated [16].
Analysis of molecular variance and F-statistics
The genotypic data were subjected to various measures of the genetic relationships within and among the durum wheat genotypes using GeneAlex version: 6.5 [17]. Genetic parameters such as total number of alleles per locus (Na), number of effective alleles per locus (Ne), Shannon's Information Index (I), and gene diversity were determined according to Nei [18].
The F-statistics such as genetic differentiation (FST), fixation index or inbreeding coefficient (FIS), and overall fixation index (FIT) were calculated according to Wright's original derivation [19]. The non-parametric Analysis of Molecular Variance (AMOVA) was conducted by using Gene Alex version: 6.5 software package based on Euclidian distance [20]. Gene flow (Nm) was calculated as: Nm=1/4*[(1-FST)/ FST)] where FST is genetic differentiation [21]. Polymorphic information content (PIC) was estimated using Power marker version: 3.25.
Genetic distance and cluster analysis
To examine the degree of population differentiation among the study materials, the Nei’s unbiased genetic distance and genetic identity were computed according to Nei [18] using GeneAlex version: 6.5 software package. The neighbour joining (NJ) method, Saitou et al. [22] and Studier et al. [23] was used to compare individual genotypes and evaluate patterns of genotypic clustering.
Results
Analysis of molecular variance (AMOVA)
Analysis of molecular variance (AMOVA) showed a lower proportion of variation among populations (19%) as compared to variation within population (81%) (Table 3).
| Sources of Variation | df | SS | MS | Estimated (%) | Variation | Statistics | Values | P | F-statistics |
|---|---|---|---|---|---|---|---|---|---|
| Among populations (AP) | 14 | 289.6 | 20.69 | 1.398 | 0.19 | - | - | - | Fis=0.398 |
| Within population (WP) | 145 | 849.36 | 5.86 | 5.858 | 0.81 | PhiPT | 0.2 | 0 | Fit=0.26 |
| Total (TOT) | 159 | 1139 | - | 7.255 | 1 | - | - | - | Fst=0.10 |
*df stands for degrees of freedom; SS for sum of square; MS for mean squares; PhiPT=AP/(WP+AP)=AP/TOT, Fis: Inbreeding coefficient; Fit: overall fixation index; Fst: Genetic differentiation.
Table 3: Analysis of molecular variance (AMOVA) showing the distribution of genetic diversity within and among populations of durum wheat genotypes from different sources of origins.
The F-statistics values across each locus and over all loci were computed as summarized in (Table 3). It was found that all loci exhibited low level of inbreeding (Fis=0.398). The mean genetic differentiation estimated by F-statistics at all loci was moderately high (Fst=0.10), indicating existence of high levels of variation among the individuals. The mean gene flow of 12 loci for the tested 160 genotypes was moderate (Fit=0.26).
Magnitude of genetic diversity
Genetic distances: The genetic distances (GD) among the genotypes ranged from 0.092 to 0.576 (Table 4). The distance between improved genotypes released from the national durum wheat improvement program and the landrace collections ranged from 0.245 to 0.518. Materials collected from Eastern Tigray were found to be the most divergent from those collected from the rest of the sources (GD=0.212-0.576), which could be explained by geographic isolation of the region. Collections from North Gonder and East Tigray were the most divergent (GD=0.576), followed by collections from Bale and Arsi (GD=0.550) and between the improved varieties and accessions of South Wello (GD=0.528. This could be due to low level of gene exchange because of geographic distance between those areas or due to wide genetic differentiation between accessions of these areas. The smallest genetic distance (GD=0.092) was observed between accessions from Bale and Harergie.
| Geographical Origin of Region | SW | SG | WG | EG | NG | NS | Arsi | Bale | HAR | ES | WS | ST | CT | ET | RV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| South Wello (SW) | 0 | ||||||||||||||
| South Gonder (SG) | 0.112 | 0 | |||||||||||||
| West Gojam (WG) | 0.448 | 0.469 | 0 | ||||||||||||
| East Gojam (EG) | 0.274 | 0.252 | 0.157 | 0 | |||||||||||
| NorthGonder (NG) | 0.19 | 0.18 | 0.317 | 0.142 | 0 | ||||||||||
| North Shewa (NS) | 0.279 | 0.22 | 0.517 | 0.376 | 0.398 | 0 | |||||||||
| Arsi | 0.456 | 0.306 | 0.475 | 0.351 | 0.528 | 0.154 | 0 | ||||||||
| Bale | 0.487 | 0.406 | 0.454 | 0.48 | 0.55 | 0.48 | 0.359 | 0 | |||||||
| Harergie (HAR) | 0.413 | 0.322 | 0.341 | 0.365 | 0.426 | 0.365 | 0.271 | 0.092 | 0 | ||||||
| East Shewa (ES) | 0.217 | 0.228 | 0.426 | 0.442 | 0.304 | 0.192 | 0.406 | 0.483 | 0.37 | 0 | |||||
| West Shewa (WS) | 0.404 | 0.363 | 0.484 | 0.488 | 0.439 | 0.392 | 0.451 | 0.382 | 0.235 | 0.246 | 0 | ||||
| Southern Tigray (ST) | 0.358 | 0.268 | 0.41 | 0.381 | 0.343 | 0.211 | 0.276 | 0.305 | 0.246 | 0.198 | 0.127 | 0 | |||
| Central Tigray (CT) | 0.329 | 0.24 | 0.396 | 0.427 | 0.323 | 0.328 | 0.432 | 0.492 | 0.391 | 0.257 | 0.248 | 0.122 | 0 | ||
| Eastern Tigray (ET) | 0.467 | 0.384 | 0.475 | 0.513 | 0.576 | 0.276 | 0.212 | 0.349 | 0.361 | 0.46 | 0.5 | 0.276 | 0.3 | 0 | |
| Released variety (RV) | 0.518 | 0.407 | 0.289 | 0.307 | 0.245 | 0.394 | 0.497 | 0.457 | 0.409 | 0.405 | 0.418 | 0.282 | 0.275 | 0.314 | 0 |
Table 4: Geographic isolation of the regions.
Pattern of genetic diversity
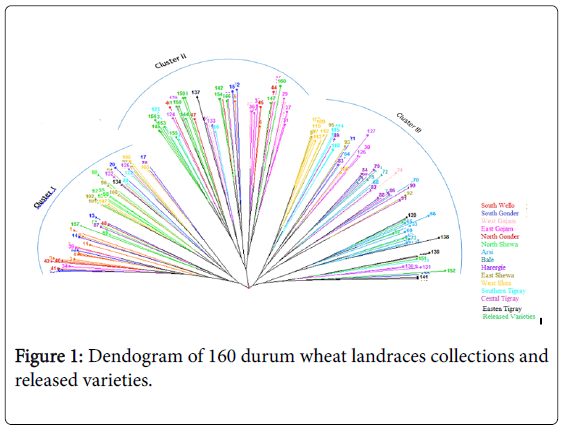
The dendogram based on neighbor-joining algorithm categorized the 160 durum wheat genotypes into three major clusters. Cluster I consisted of 47 genotypes, cluster II 64 genotypes and cluster III 49 genotypes (Figure 1).
The cross-border similarities between a few adjoining regions may be attributed, at least in part, to seed movements among neighboring regions. The first cluster consisted most of the genotypes collected from South Wello, South Gonder, North and East Shewa. Most of the released varieties and half of the landraces of north Gonder and most of those from East Gojam were grouped to cluster II. On the other hand, Cluster III consisted most of the genotypes collected from Harergie, West Shewa, Central and Eastern Tigray and all of the landraces collected from Arsi and Bale (Table 5).
| Geographical Region of Origin | Number of Genotypes | Clusters | ||
|---|---|---|---|---|
| C I | C II | C III | ||
| South Wello | 11 | 0.819 | 0.182 | 0 |
| South Gonder | 11 | 0.637 | 0.182 | 0.182 |
| West Gojam | 6 | 0 | 0.833 | 0.167 |
| East Gojam | 11 | 0.182 | 0.727 | 0.091 |
| North Gonder | 9 | 0.444 | 0.556 | 0 |
| North Shewa | 11 | 0.727 | 0 | 0.243 |
| Arsi | 11 | 0 | 0 | 1 |
| Bale | 7 | 0 | 0 | 1 |
| Harergie | 14 | 0.143 | 0.071 | 0.785 |
| East Shewa | 11 | 0.636 | 0 | 0.364 |
| West Shewa | 11 | 0.364 | 0 | 0.636 |
| Southern Tigray | 10 | 0.4 | 0.4 | 0.4 |
| Central Tigray | 10 | 0.2 | 0.3 | 0.5 |
| Eastern Tigray | 8 | 0.125 | 0.125 | 0.167 |
| Released varieties | 19 | 0.052 | 0.842 | 0.105 |
Table 5: Estimated probabilistic memberships of durum wheat populations from different geographic origins to the 3 clusters.
It appears that landraces collected from South Wello share a portion of their ancestral gene pool with landraces collected from North Gonder (Figure 1). Accessions collected from West Shewa shared some part of ancestral gene pool with the accessions from the adjoining regions of East Shewa on one side and with Harergie on the other side. Accessions collected from South Wello, East Gojam, Harergie, and East and West Shewa were distributed over the three clusters, as also partly confirmed by Shannon index. This indicated that there was high genetic diversity between genotypes collected from these areas.
In contrary, genotypes collected from Arsi and Bale areas were grouped only to cluster III. The genotypes of the two areas exhibited low genetic variability may be because they share similar ancestral gene pool. Moreover, the prior introduction of high yielding and dwarf bread wheat varieties to Bale and Arsi might have contributed towards narrowing genetic base in this region.
Most of the improved varieties, grouped to cluster II, shared similar ancestral gene pool with the accessions collected from West Gojam, East Gojam and North Gonder. This might be due to the dissemination of similar released varieties to farmers of these areas and gene-flow to landraces. The genetic variability of landraces collected from South Wello and East Gojam were lower than that of landraces from Harergie and most of Tigray.
Genetic polymorphism of SSR markers
The difference between the longest and shortest amplified fragment size ranged from 100 to 350 bp, the highest size being that of primer CFD 257 (250-350 bp) and the lowest being that of primer CFA 2278 (100-180 bp). All the 12 SSR primer pairs were polymorphic and generated a total of 74 alleles (Table 6).
| Locus | Genetic Parameters* | |||||||
|---|---|---|---|---|---|---|---|---|
| Na | Ne | I | Ho | He | uHe | F | PIC | |
| BARC153 | 6 | 4.09 | 1.49 | 0.82 | 0.74 | 0.78 | -0.35 | 0.75 |
| CFA2278 | 7 | 3.72 | 1.38 | 0.97 | 0.72 | 0.76 | -0.38 | 0.82 |
| CFD 257 | 5 | 3.57 | 1.32 | 0.78 | 0.7 | 0.74 | -0.46 | 0.74 |
| WMS53 | 9 | 4.12 | 1.49 | 0.8 | 0.75 | 0.78 | -0.35 | 0.87 |
| WMS120 | 6 | 4.05 | 1.49 | 0.81 | 0.74 | 0.78 | -0.36 | 0.78 |
| WMS234 | 8 | 4.46 | 1.58 | 0.83 | 0.77 | 0.81 | -0.3 | 0.85 |
| WMS269 | 7 | 4.3 | 1.55 | 0.77 | 0.76 | 0.8 | -0.31 | 0.89 |
| WMS375 | 5 | 3.59 | 1.35 | 0.78 | 0.71 | 0.75 | -0.42 | 0.74 |
| WMS493 | 5 | 2.78 | 1.1 | 0.73 | 0.62 | 0.66 | -0.63 | 0.68 |
| WMS516 | 6 | 3.4 | 1.3 | 0.79 | 0.69 | 0.73 | -0.46 | 0.76 |
| WMS 532 | 6 | 3.6 | 1.37 | 0.81 | 0.7 | 0.74 | -0.44 | 0.78 |
| XGWM136 | 6 | 3.42 | 1.3 | 0.77 | 0.69 | 0.73 | -0.45 | 0.72 |
| Mean | 6 | 3.76 | 1.39 | 0.79 | 0.72 | 0.76 | -0.41 | 0.76 |
| SE | 0.54 | 0.45 | 0.13 | 0 | 0.04 | 0.04 | 0.09 | 0.22 |
*Nm: gene flow; Na: number of alleles; Ne: number of effective alleles; I: Shannon’s information index; Ho: observed heterozygosity; He: unbiased expected heterozygosity; F: fixation index; Nm: gene flow; PIC: polymorphic information content; and SE: standard error.
Table 6: Summary of genetic parameters for Durum wheat landraces, wild relatives and released varieties using 12 selected SSR markers.
The number of alleles per locus ranged from 4 for primers WMS 375, WMS 493, and CFD 257 to 9 for primer WMS 53, the average number of alleles per locus being six. The number of effective alleles (Ne) ranged from 3.63 for primer WMS 493 to 5.75 for WMS 234, with a mean of 4.81 (Table 6).
The observed heterozygosity (Ho) varied from 0.99 for primer CFA 2278 to 1.00 for the entire remaining locus, the mean Ho being 0.99. Gene diversity (He) of the primers of the marker ranged from 0.62 to 0.77, primer WMS 493 with the least score and primer WMS 234 with the highest score the mean He score being 0.72. The results obtained from the present study using 160 durum wheat genotypes and 12 SSR primer pairs, revealed 74 alleles with an average of six alleles per locus. The high heterozygosity and low genetic fixation values from the current study may a signal for the existence of high genetic variability within the sampled durum wheat genotypes. Some loci were found to be more mono-morphic (WMS 493, WMS 516, and WMS 532) with less gene flow than the others (e.g., WMS 269 and WMS 234) (Table 6).
The genetic diversity contribution of the SSR primers was grouped based on the polymorphic information content (PIC), using the criteria of Vaiman et al. [24] which considers loci polymorphism (PIC) value of more than 0.5 as high, PIC value between 0.5 and 0.25 as medium and PIC value less than 0.25 as low. The polymorphic information content (PIC) of the loci ranged from 0.69 (WMS 493) to 0.89 (WMS 269), with a mean of 0.76 (Table 6). It was found that the entire 12 loci used in the present study were highly informative throughout the genome of 160 durum wheat genotypes. The numbers of observed and effective alleles were higher for the landraces than improved varieties. Relatively the highest Shannon diversity index was recorded for the landraces collected from south Gonder (1.73), while the population collected from Bale (1.17) showed the least variability. The mean value of Shannon diversity index was 1.39 (Table 7). The gene diversity (observed hetrozygosity) of genotypes included in this study ranged from 0.79 (West Shewa) to 0.99 (East Gojam), with the mean value of 0.92 (Table 7).
| Geographical Region of Origin | N | Na | Ne | I | Ho | He | uHe | F | P% |
|---|---|---|---|---|---|---|---|---|---|
| South Wello | 11 | 5.92+0.313 | 4.66+0.260 | 1.38+0.078 | 0.92+0.00 | 0.71+0.024 | 0.74+0.025 | -0.37 | 91.63% |
| South Gonder | 11 | 6.5+0.195 | 5.29+0.26 | 1.73+0.054 | 0.94+0.00 | 0.76+0.016 | 0.79+0.017 | -0.31 | 98.65% |
| West Gojam | 6 | 5.25+0.329 | 4.53+0.24 | 1.31+0.079 | 0.96+0.00 | 0.69+0.024 | 0.76+0.026 | -0.39 | 92.67% |
| East Gojam | 11 | 6.17+0.207 | 5.06+0.313 | 1.46+0.072 | 0.99+0.00 | 0.73+0.023 | 0.77+0.025 | -0.33 | 98.53% |
| North Gonder | 9 | 6.3+0.188 | 5.01+0.28 | 1.49+0.061 | 0.91+0.00 | 0.74+0.019 | 0.78+0.021 | -0.32 | 91.67% |
| North Shewa | 11 | 5.92+0.336 | 4.69+0.253 | 1.38+0.076 | 0.89+0.00 | 0.71+0.024 | 0.75+0.025 | -0.37 | 90.70% |
| Arsi | 11 | 6.08+0.29 | 4.59+0.29 | 1.36+0.076 | 0.93+0.00 | 0.70+0.024 | 0.73+0.025 | -0.4 | 83.33% |
| Bale | 7 | 4.92+0.39 | 3.13+026 | 1.178+0.095 | 0.94+0.00 | 0.65+0.029 | 0.70+0.032 | -0.49 | 75.00% |
| Harergie | 14 | 6.83+0.207 | 5.59+0.218 | 1.37+0.055 | 0.97+0.00 | 0.71+0.018 | 0.74+0.019 | -0.38 | 98.86% |
| East Shewa | 11 | 6.17+0.207 | 5.82+0.248 | 1.43+0.059 | 0.95+0.00 | 0.72+0.019 | 0.76+0.020 | -0.35 | 75.00% |
| West Shewa | 11 | 5.33+0.310 | 4.44+0.224 | 1.29+0.079 | 0.79+0.01 | 0.69+0.026 | 0.72+0.027 | -0.39 | 75.00% |
| Southern T. | 10 | 6.08+0.193 | 5.07+0.205 | 1.47+0.051 | 0.90+0.00 | 0.75+0.016 | 0.78+0.017 | -0.32 | 90.67% |
| Central T. | 10 | 6.5+0.174 | 4.79+0.139 | 1.43+0.028 | 0.94+0.00 | 0.73+0.010 | 0.77+0.011 | -0.35 | 91.65% |
| Eastern T. | 8 | 5.75+0.329 | 4.60+0.252 | 1.36+0.075 | 0.80+0.00 | 0.71+0.022 | 0.75+0.023 | -0.383 | 74.00% |
| Released V. | 19 | 5.33+0.376 | 5.01+0.239 | 1.47+0.056 | 0.93+0.00 | 0.74+0.014 | 0.76+0.015 | -0.324 | 91.79% |
| Mean+ SE | 11 | 5.91+0.076 | 4.76+0.07 | 1.39+0.02 | 0.92+0.01 | 0.72+0.006 | 0.75+0.006 | -0.398 | 88+2.41 |
Na: number of alleles; Ne: number of effective alleles; I: Shannon’s information index; Ho: observed heterozygosity; He: unbiased expected heterozygosity; F: fixation index; and SE: standard error.
Table 7: Mean of genetic parameters for sixteen groups of the 160 durum wheat genotypes studied using 12 selected SSR markers.
The degree of polymorphism between populations varied from 74% for the collections from Eastern Tigray to 98.86% for collections from Harergie, with the average of 88%. Landraces collected from Harergie, South Gonder and East Gojam exhibited 98.86%, 98.65% and 98.53% polymorphism, respectively. This is higher than the values scored from the rest of the collections from other origins. Relatively lower amounts of polymorphism were observed from the landraces collected from Eastern Tigray (74%), East and West Shewa (75%) and Bale (75%) zones. The remaining collection areas showed moderate polymorphism that ranged from 83-92%. The improved varieties also showed a polymorphism value of 91.67%, which is higher than polymorphism values scored by some of the local collections (Table 6).
Discussion
Analysis of molecular variance
Analysis of molecular variance partitioned the total genetic variance into variance among populations of 19% and within population of 81%. This result agreed with a number of reports from previous studies [25,26]. The low genetic differentiation observed between populations may be due to migration or selection by farmers for similar traits. AMOVA was used to determine that variation among and within groups was highly significant (p<0.001), with the clusters capturing 31.5% of the total genetic variations, while 68.3% was explained by individuals within populations [27].
All locus exhibited low level of inbreeding (0.39), which resulted in homozygosity, indicating increase the chances of offspring being affected by recessive or deleterious traits. Low inbreeding value of all loci for the tested durum wheat genotypes indicated existence of wider genetic distance between the genotypes or lower effects of non-random mating. Fixation index (FST) is a measure of population differentiation due to genetic structure. The mean genetic differentiation estimated by F-statistics at all loci was moderately high (0.10) which could be due to high levels of variation among individuals [28].
Magnitude of genetic diversity
From Nie’s pairwise genetic distance, the highest genetic distance was observed between accessions from North Gonder and East Tigray (0.57). Crossing genotypes from genetically distant clusters may result in the expression of more heterosis in F1 generation and wider genetic variability in the segregating generations. Parents for hybridization could be selected on the basis of large inter-cluster distances for isolating useful recombinants in the segregating generations [29]. On the other hand, the minimum genetic distance was observed between accessions collected from Bale and Hararge (0.092). This might be due to high seed exchange between these areas, as the areas were under a single administrative zone in the past and this might be creating high chance of seed exchange between farmers. The differences observed in landrace populations studied could also result at least partly from combined effects of genetic drift, mutation, migration and selection [30]. The cross-border similarities between a few adjoining regions may be attributed, at least in part, to seed movements among neighboring regions [31].
Patterns of genetic diversity
The populations collected from 14 zones of three regions (Tigray, Amhara and Oromia) and released varieties were grouped into 3 clusters of distinct genetic background. These grouping of genotypes into distinct clusters showed that they had evolved from different lines of ancestry or derived from independent events of evolutionary forces (genetic drift, mutation, migration, selection and in flux/out flux of genes in the form of germplasm exchange) that separated them into different gene pools [32]. The clustering pattern did not clearly show the existence of definite pattern of relationships between geographical origins and genetic diversity for microsatellite markers. Populations from the same geographical origin were found to fall into single or different clusters. Similarly, genotypes from different origins were also grouped into a single cluster or in different clusters. Some clusters contained populations from the same geographical origin while others had populations from different geographical origins and the number of genotypes varied from cluster to cluster. Similar results were recorded for both the hexaploid and tetraploid wheats in another study [33].
A number of authors also found similar results. Bousba et al. [13], for instance, observed no definite relationship between sources of geographic origin and genetic diversity in durum wheat collections from different countries. Similar result was found from the study on chickpea germplasms collected from Ethiopia [32]. Ijaz et al. [34] reported the same scenario after their study of 48 accessions and 15 varieties that the cluster analysis separated the accessions and the landraces in to two distinct groups [35]. Similarly, Jemanesh et al. also evaluated 58 tetraploid wheat accessions including landraces and advanced improved varieties by 31 neutral SSR markers and observed low variability among the released cultivars. Moreover, seed exchange among neighboring durum wheat growing regions could also contribute to the observed higher within population variation. This result was in accordance with the results of the previous studies [14,36,37].
The landraces and improved varieties were grouped into three clusters, with all landraces being distributed over all clusters. This might be due to the fact that released varieties and landraces may share common ancestor. The landraces are grouped together with improved varieties and there is no definite distinction between them as opposed to previous studies. Mondal et al. [38] reported that improved durum wheat varieties from Ethiopia were clustered into the same group except for the landrace variety ‘DZ04’ which was clustered with the landraces. Tesfaye et al. [39], also assessed the genetic diversity of these wheat varieties based on gliadin alleles and found the distinct clustering of landraces with improved varieties. Similar scenario was observed from a study by Ijaz et al. [34], who observed two distinct groups of landraces and improved varieties. Altintas et al. [35], reported that ‘Kunduru’, a durum wheat variety that was directly selected from a landrace, was the most distinct variety in their study compared to the other Turkish durum wheat varieties that were derived from CIMMYT germplasm. Similarly, in our study some varieties that were selected from Ethiopian landraces were more closely related to the landraces than the varieties. Zarkti et al. [40], indicated that the extent of genetic diversity in genotypes selected from local germplasm was high.
Genetic polymorphism of SSR markers
In this study, we assessed genetic diversity of Ethiopian durum wheat landraces collected from the three major wheat producing regions (Amhara, Oromia and Tigray) and improved varieties released in Ethiopia using 12 SSR primers. The average number of alleles per locus of 74 alleles (an average of 6 alleles per locus) we observed in this study is relatively lower than the number recorded in a number of previous studies. For instance, Eujayl et al. [36] from an assessment of genetic diversity in 58 tetraploid wheat accessions including landraces and advanced improved varieties using 31 neutral SSR primer pairs, identified a total of 286 alleles, with 9.2 alleles per locus. Teklu et al. [41] examined 141 Ethiopian tetraploid wheat landraces consisting of three species (Triticum durum Desf., T. dicoccon Schrank and T. turgidum L.) using 29 microsatellite markers and identified 320 alleles (an average number of 11.3). Similar results were also obtained from a number of similar studies in Ethiopia [33] and elsewhere [3,42]. The lower the number of alleles found from the present study might be due to the reduction of the genetic diversity between landraces because of the recently encountered higher varietal replacement rates of durum with bead wheat.
Four microsatellites (CFA 2278, WMS 53, WMS 234, and WMS 269), each with more than 6 alleles per loci, were found to be the most polymorphic, indicating their better discrimination ability and usefulness for further genetic diversity studies and marker-assisted selection in durum wheat. Similar results on polymorphic information content were observed from a previous study durum wheat genotypes using SSR marker [13]. High heterozygosity and low genetic fixation values were observed from this study including in released varieties, indicating that the durum wheat breeding program in Ethiopia has been utilizing diverse source of genes to develop new varieties. The present results are also in agreement with the high genetic variability of durum wheat reported by Bousba et al. [13].
The populations collected from 14 zones of three regions (Tigray, Amhara and Oromia) and released varieties were grouped into 3 clusters of distinct genetic background. These grouping of genotypes in to different distinct clusters showed that they had evolved from different lines of ancestry or derived from independent events of evolutionary forces (genetic drift, mutation, migration, selection and in flux/out flux of genes in the form of germplasm exchange) that separated them into related but different gene pools [32]. The clustering pattern did not clearly show the existence of definite pattern of relationships between geographical origins and genetic diversity for microsatellite markers. Populations from the same geographical origin were found to fall into single or different clusters. Similarly, genotypes from different origins were also grouped into a single cluster or in different cluster. Some clusters contained populations from the same geographical origin while others had populations from different geographical origins and the number of genotypes varied from cluster to cluster. Similar results were recorded for both the hexaploid and tetraploid wheat in another study [33].
Conclusion
The high genetic variation detected in this study shows the existence of ample variability that can be exploited for future durum wheat breeding program as a source of valuable traits. Furthermore, the high variability noted within the tested improved varieties (0.93) indicated that the durum wheat breeding program in Ethiopia has been utilizing diverse source of genes to develop new varieties. Based on the result of this study, it is to conclude that the genetic diversity of Ethiopian durum wheat gradually reduced compared with the previous reports. The cause of reduction of the genetic diversity might be due to the high rate of replacement durum wheat by bread wheat, high gene flow between adjoining zones and may be duplication of collections of similar genotypes with different accession number.
References
- Friebe B (1996) Chromosome banding and genome analysis in diploid and cultivated polyploid wheats. Methods of Genome Analysis in Plants, pp: 39-60.
- Von Büren M (2001) Polymorphisms in two homeologous γ-gliadin genes and the evolution of cultivated wheat. Genet Resour Crop Evol 48: 205-220.
- Colomba MS, Gregorini A (2011) Genetic diversity analysis of the durum wheat Graziella Ra, Triticum turgidum L. subsp. durum (Desf.) Husn.(Poales, Poaceae). Biodivers J 2: 73-84.
- Singh BD (2002) Plant breeding: principles and methods. Kalyani, New Delhi.
- Chahal GS, Gosal SS (2002) Principles and procedures of plant breeding: Biotechnological and conventional approaches. Alpha Science Int'l Ltd.
- Gebre-Mariam H (1991) Wheat production and research in Ethiopia. Wheat Research in Ethiopia: A Historical Perspective, pp: 1-16.
- Uddin MS, Boerner A (2008) Genetic diversity in hexaploid and tetraploid wheat genotypes using microsatellite markers. Plant Tissue Cult Biotech 18: 65-73.
- Jain SK, Qualset CO, Bhatt GM, Wu KK (1975) Geographical patterns of phenotypic diversity in a world collection of durum wheats 1. Crop Sci 15: 700-704.
- Bekele E (1984) Analysis of regional patterns of phenotypic diversity in the Ethiopian tetraploid and hexaploid wheats. Hereditas 100: 131-154.
- Negassa M (1986) Patterns of diversity of Ethiopian wheats (Triticum spp.) and a gene center for quality breeding. Plant Breed 97: 147-162.
- Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C, et al. (2003) Noncoding plastid trnT‐trnF sequences reveal a well resolved phylogeny of basal angiosperms. Journal of evolutionary Biology 16: 558-576.
- Mondini L, Farina A, Porceddu E, Pagnotta MA (2008) Assessment of the genetic variation in Ethiopian germplasm of durum wheat from region with contrasting environment and water stress. In 11th International Wheat Genetics Symposium 2008 Proceedings 1: 261.
- Bousba R, Baum M, Djekoune A, Labadidi S, Djighly A (2012) Screening for drought tolerance using molecular markers and phenotypic diversity in durum wheat genotypes. World Appl Sci J 16: 1219-1226.
- Abouzied HM, Eldemery SM, Abdellatif KF (2013) SSR-based genetic diversity assessment in tetraploid and hexaploid wheat populations. Br Biotechnol J 3: 390.
- Seetharam K, Thirumeni S, Paramasivam K (2009) Estimation of genetic diversity in rice (Oryza sativa L.) genotypes using SSR markers and morphological characters. Afr J Biotechnol 8: 2050-2059.
- Sarla N, Neeraja CN, Siddiq EA (2005) Use of anchored (AG)n and (GA)n primers to assess genetic diversity of Indian landraces and varieties of rice. Current Sci, pp: 1371-1381.
- Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288-295.
- Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genet 89: 583-590.
- Wright S (1949) The genetical structure of populations. Annals of Eugenics 15: 323-354.
- Ge S, Oliveira GC, Schaal BA, Gao LZ, Hong DY (1999) RAPD variation within and between natural populations of the wild rice Oryza rufipogon from China and Brazil. Heredity 82: 638.
- Slatkin M, Barton NH (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349-1368.
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406-425.
- Studier JA, Keppler KJ (1988) A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol 5: 729-731.
- Vaiman D, Mercier D, Moazami-Goudarzi K, Eggen A, Ciampolini R, et al. (1994) A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mammalian Genome 5: 288-297.
- Teklu Y, Hammer K (2009) Diversity of Ethiopian tetraploid wheat germplasm: breeding opportunities for improving grain yield potential and quality traits. Plant Genet Resour 7: 1-8.
- Alayachew SA, Geletu KT (2017) Genetic diversity of Ethiopian emmer wheat Triticum dicoccum Schrank landraces using seed storage proteins markers. Afr J Biotechnol 16: 889-894.
- Kabbaj H, Sall AT, Al-Abdallat A, Geleta M, Amri A, et al. (2017) Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Frontiers in Plant Science 8: 1277.
- Hartl DL, Clark AG (1989) Principles of population genetics. Sinauer Associates. Sunderland, MA.
- Ghaderi A, Adams MW, Nassib AM (1984) Relationship between genetic distance and heterosis for yield and morphological traits in dry edible bean and faba bean 1. Crop Sci 24: 37-42.
- Felsenstein J (2007) Theoretical evolutionary genetics. University of Washington, USA.
- Keneni G, Bekele E, Getu E, Imtiaz M, Dagne K, et al. (2011) Characterization of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions for response to infestation by adzuki bean beetle (Callosobruchus chinensis L.) I. Performance Evaluation. Ethiop J Agric Sci 21: 65-83.
- Keneni G, Bekele E, Assefa F, Imtiaz M, Debele T, et al. (2013) Evaluation of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions for symbio-agronomic performance. Renewable Agriculture and Food Systems 28: 338-349.
- Alamerew S, Chebotar S, Huang X, Röder M, Börner A (2004) Genetic diversity in Ethiopian hexaploid and tetraploid wheat germplasm assessed by microsatellite markers. Genet Res Crop Evol 51: 559-567.
- Ijaz S, Khan IA (2009) Molecular characterization of wheat germplasm using microsatellite markers. Genet Mol Res 8: 809-815.
- Altıntaş S, Toklu F, Kafkas S, Kilian B, Brandolini A, et al. (2008) Estimating genetic diversity in durum and bread wheat cultivars from Turkey using AFLP and SAMPL markers. Plant Breeding 127: 9-14.
- Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor App Genet 104: 399-407.
- Jemanesh KH, Karl-Hammer AB, Miloudi M, Nachit MS, Ro¨der KO, et al. (2010) Genetic diversity assessment of Ethiopian tetraploid wheat landraces and improved durum wheat varieties using microsatellites and markers linked with stem rust resistance exploiting a wheat EST database to assess genetic diversity. Genet Mol Biol 33: 719-730.
- Mondal MAA (2003) Improvement of potato (Solanum tuberosum L.) through hybridization and in vitro culture technique. Rajshahi University, Rajshahi, Bangladesh.
- Tesfaye K, Govers K, Bekele E, Borsch T (2005) ISSR fingerprinting of wild coffea arabica in Ethiopia reveals high levels of genetic diversity within regions. In BioTeam Status seminar, Bonn, Germany.
- Zarkti H, Ouabbou H, Hilali A, Udupa SM (2010) Detection of genetic diversity in Moroccan durum wheat accessions using agro-morphological traits and microsatellite markers. Afr J Agric Res 5: 1837-1844.
- Teklu Y, Hammer K (2006) Farmers’ perception and genetic erosion of tetraploid wheats landraces in Ethiopia. Genet Res Crop Evol 53: 1099-1113.
- Huang X, Börner A, Röder M, Ganal M (2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor App Genet 105: 699-707.
Citation: Asmamaw M, Keneni G, Tesfaye K (2019) Genetic Diversity of Ethiopian Durum Wheat (Triticum durum Desf.) Landrace Collections as Reveled by SSR Markers. Adv Crop Sci Tech 7: 413. DOI: 10.4172/2329-8863.1000413
Copyright: © 2019 Asmamaw M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 3723
- [From(publication date): 0-2019 - Apr 25, 2024]
- Breakdown by view type
- HTML page views: 2939
- PDF downloads: 784