Published online Aug 26, 2015. doi: 10.4331/wjbc.v6.i3.110
Peer-review started: March 28, 2015
First decision: April 27, 2015
Revised: May 8, 2015
Accepted: July 16, 2015
Article in press: July 17, 2015
Published online: August 26, 2015
Hyaluronan is a rapidly turned over component of the vertebrate extracellular matrix. Its levels are determined, in part, by the hyaluronan synthases, HAS1, HAS2, and HAS3, and three hyaluronidases, HYAL1, HYAL2 and HYAL3. Hyaluronan binding proteins also regulate hyaluronan levels although their involvement is less well understood. To date, two genetic disorders of hyaluronan metabolism have been reported in humans: HYAL1 deficiency (Mucopolysaccharidosis IX) in four individuals with joint pathology as the predominant phenotypic finding and HAS2 deficiency in a single person having cardiac pathology. However, inherited disorders and induced mutations affecting hyaluronan metabolism have been characterized in other species. Overproduction of hyaluronan by HAS2 results in skin folding and thickening in shar-pei dogs and the naked mole rat, whereas a complete deficiency of HAS2 causes embryonic lethality in mice due to cardiac defects. Deficiencies of murine HAS1 and HAS3 result in a predisposition to seizures. Like humans, mice with HYAL1 deficiency exhibit joint pathology. Mice lacking HYAL2 have variably penetrant developmental defects, including skeletal and cardiac anomalies. Thus, based on mutant animal models, a partial deficiency of HAS2 or HYAL2 might be compatible with survival in humans, while complete deficiencies of HAS1, HAS3, and HYAL3 may yet be recognized.
Core tip: This manuscript summarizes the phenotypes that have been associated with alterations in hyaluronan synthesis or degradation. It should serve as a reference for those who are considering an alteration in hyaluronan metabolism as the cause of a genetic condition.
- Citation: Triggs-Raine B, Natowicz MR. Biology of hyaluronan: Insights from genetic disorders of hyaluronan metabolism. World J Biol Chem 2015; 6(3): 110-120
- URL: https://www.wjgnet.com/1949-8454/full/v6/i3/110.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i3.110
Hyaluronan (HA) is a straight chain polymer comprised of repeating disaccharide units of glucuronic acid and N-acetylglucosamine (→4]β-D-GlcA-[1→3]-β-D-GlcNAc-[1→)n, and has substantial size heterogeneity in different tissues, although in most tissues it is greater than 106 daltons[1]. Although the presence of both uronic acid and an amino sugar define HA as a glycosaminoglycan (previously known as mucopolysaccharides), it differs from the other members of this family of compounds in that it is not sulfated or protein-linked, and rather than being synthesized in the Golgi, it is synthesized at the cell membrane and extruded into the extracellular space[2]. HA can assemble with tissue-specific binding proteins known as hyaladherins to form large complexes. Within the extracellular matrix, many structural and biological functions have been attributed to HA of different sizes. Examination of the phenotypes associated with naturally-occurring and laboratory-induced alterations in HA metabolism has facilitated knowledge of HA metabolism and function.
It is estimated that HA accounts for 15 g of the “average” 70 kg human[3]. Studies of adults have revealed the highest HA concentrations in soft connective tissues such as skin, synovial fluid, Wharton’s jelly of the umbilical cord, and vitreous body[4]. The concentration of HA is also very high during embryogenesis and wound healing[5]. Low concentrations of HA are present in serum, where there is an average of 20-40 μg per liter[6,7], although there are many pathological states that are associated with and may cause increased levels of serum HA[4,8]. Based on whole body studies in the rat and other species, it is estimated that 50% of the HA is in the skin, 25% in the skeleton and joints, and most of the remaining HA is in brain, kidney, lung and muscles[4,9].
HA is critical to the assembly and structure of the extracellular matrix at both tissue and cellular levels. The large size of HA provides rigidity to some types of matrix and serves a scaffolding function for several constituents of the matrix. The hydrophilic groups of HA attract water to hydrate tissues, lubricate joints, and fill space. A HA-enriched, hydrated matrix facilitates cellular movement and proliferation that is critical for cellular migrations during early developmental processes and in the regeneration and remodeling of tissues such as during wound healing. HA has also been shown to play a role in inflammation and ovulation[10,11]. The functions of HA depend on its size; high molecular mass and low molecular mass HA typically have opposing effects on epithelial to mesenchymal transition, inflammation, and angiogenesis[12,13].
Among extracellular matrix molecules, HA has one of the fastest rates of turnover[1,14,15], with an estimated one-third of the 15 g of HA in an average adult human turned over daily[8]. The half-life of HA was initially estimated to be 2-4 d[15], but it is now clear that the half-life varies among and within tissues because of differences in the rates of degradation or other forms of turnover. For example, HA has a half-life of about 2-5 min in blood[16,17], whereas it is estimated to be 2.5 d in the skin[18], and up to 18 d when bound to aggrecan in cartilage[19]. Approximately 30% of HA degradation occurs locally, within the tissue in which it is synthesized[18]. The remaining about 70% enters the lymphatic drainage where 90% is removed by the lymph nodes[20] and the remainder is taken up from the blood by the liver[16] and, to a lesser extent, the kidney and spleen[21]. Sinusoidal endothelial cells are responsible for uptake by the liver[22-24] via receptors with high affinity for HA and chondroitin sulfate[25]. Only about 1% of blood HA is eliminated each day in the urine[26], and this is limited to lower molecular mass HA (< 12000 Da) that can pass through the glomerular filtration barrier.
The internalization of HA is essential for its complete breakdown in cells, but identifying the receptor(s) responsible for uptake in each tissue has been challenging. Three receptors, HA receptor for endocytosis (HARE, also known as stabilin-2), cluster of differentiation antigen 44 (CD44), and lymphatic vessel endothelial receptor-1 (LYVE-1), are able to internalize HA for degradation in vitro, but their relevance in vivo is less clear. HARE localizes to endothelial cells of organs that internalize circulating HA including liver, lymph nodes and spleen, as well as to endothelial cells of the oviduct, corneal and lens epithelium, mesenchymal cells of the heart valve, ependymal cells of the brain, macrophages, and epithelial cells of the renal papillae[27]. HA uptake via clathrin-coated pits of liver endothelial cells[28] is inhibited by a HARE blocking antibody[29,30], and an anti-cluster of differentiation associated antigen 44 (CD44) blocking antibody inhibits both endocytosis and cleavage of HA[31]. The LYVE-1 receptor binds and internalizes HA in transfected human embryonic kidney cells[32]; much remains to be learned about its regulation[33]. It seems likely that each of these receptors can internalize HA under specific circumstances, but that HARE appears to be most important in the internalization of circulating HA. Only HARE knock-out mice have elevated circulating HA[30]; an increased concentration of serum HA was not found in mice lacking both the CD44 and LYVE-1 receptors[34]. CD44 appears to function primarily in the uptake of HA during pathological processes such as inflammation[35], although it may also have a role in local degradation of HA such as in cartilage[36] or developing heart[37].
Internalization of HA is likely preceded by its depolymerization. This concept was first proposed because the size of HA was found to be reduced after transit through lymph nodes[38]. In cartilage explants, a decrease in the size of steady-state HA compared to newly synthesized HA, also suggested that extracellular depolymerization of HA was an initial step in HA degradation[19]. A model whereby a cell surface hyaluronidase, putatively HYAL2 (described below), initiates the cleavage of HA before receptor-mediated internalization for intracellular degradation has been proposed[39]. Other modes of HA depolymerization, such as by oxygen-derived radicals[40], have also been suggested. The studies in cartilage explants also suggested that HA is internalized and degraded in conjunction with the binding domain of aggrecan[19], a finding that has not been explored in the context of current degradation models.
Among the HA binding proteins that have been identified, the recently described KIAA1199 is unique because its presence facilitates the degradation of HA[41]. Unlike CD44 which is concentrated in calveolin-rich lipid rafts, KIAA1199 appears to engage HA via a clathrin-coated pit pathway[41]. KIAA1199 facilitates the degradation of HA in some tissues, although it does not have homology to the active sites of mammalian hyaluronidases and it is unclear how it directly or indirectly depolymerizes HA. Variants of KIAA1199 that are associated with reduced in vitro HA degradative activity are described that have been found in several persons with non-syndromic hearing deficit[41,42]. The mouse orthologue of KIAA1199 also has HA degradative activity[43]. While the mode of action toward HA and the physiological role of KIAA1199 are not yet known, it has been proposed that KIAA1199 has a key role in HA catabolism in dermis and, possibly, brain[41].
The rapid rate of HA turnover in many tissues requires high rates of both synthesis and degradation. Taken together with the important functional roles of HA, one might expect that defects in either HA synthesis or degradation could cause clinical phenotypes. However, for these same reasons, it has been speculated that defects in HA metabolism may be lethal. To date, only two genetic disorders of HA metabolism have been described in humans and studies in the mouse provide evidence to support the earlier contentions that complete deficiencies in at least some HA metabolizing enzymes may be lethal during embryonic or fetal development.
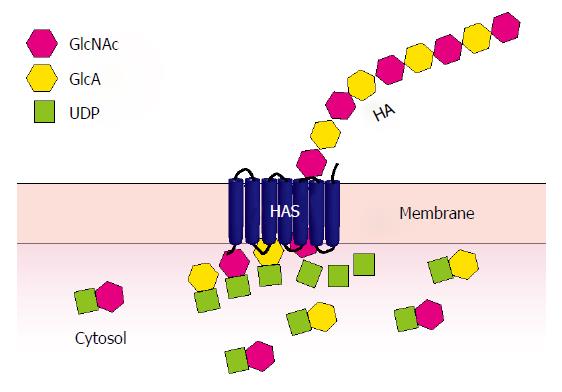
HA synthesis at the inner aspect of the plasma membrane takes place through the sequential transfer of uridine diphosphate (UDP)-GlcA and UDP-GlcNAc to the reducing terminus of the growing polysaccharide chain[44] (Figure 1). The first mammalian HA synthase to be identified, HAS1, was identified from cDNA libraries by functional expression; HAS2 and HAS3 were identified soon after by homology-based cloning[2]. Genes encoding these three enzymes, HAS1, HAS2, and HAS3, were mapped to human chromosomes 19q13.3, 8q24.12, and 16q22.1, respectively[45]. All of these enzymes are capable of HA synthesis, but of different average sizes and at different rates[46,47]. The three HAS enzymes are expressed in both embryonic and adult tissues; HAS2 is most broadly expressed during development, while HAS3 appears to be most broadly expressed in adult tissues[48,49]. The in vivo functions of the individual enzymes are becoming clearer through the characterization of mouse knockouts of these genes.
HAS1 is the least active of the synthases, requiring a very high concentration of UDP-GlcNAc[46,47]. No abnormalities were detected in initial studies of mice with a targeted disruption of Has1[50], although subsequent studies that focused on the brain revealed a mild increase in seizure activity[51]. Accelerated wound closure and decreased HA synthesis was recently demonstrated in Has1-/- Has3-/- mice, but the involvement of HAS1 appears minimal in comparison to HAS3[52]. Altered splicing of HAS1 mRNA has been reported in cases of malignant myeloma and Waldenström’s macroglobulinemia[53,54], and inherited and acquired HAS1 mutations are found in increased frequency in these conditions[55,56]. Despite this strong association, the relationship between this finding and the function of HAS1 remains to be determined.
HAS2 is recognized as the synthase responsible for the majority of HA synthesis during development[50]. Has2-/- mouse embryos die at embryonic day (E) 9.5 due to a failure to form the HA-rich cardiac cushion and initiate the epithelial to mesenchymal transition (EMT) required to form the valves and septa[57,58]. Exogenous HA could rescue the EMT defect in ex vivo explants from the developing cardiac cushion of Has2-/- embryos[58], providing evidence that HA regulates signal transduction pathways during development. The severity of the cardiac defect in Has2-/- embryos required that conditional deletions of Has2 be used to study its role in other aspects of development. A deletion of Has2 limited to the limb bud mesoderm of mice demonstrated that HA synthesis is also essential for growth and patterning of the limb, maturation of developing chondrocytes, and joint formation[59]. A nestin-cre driven deletion of Has2 did not severely impact the developing brain, although the mice had increased seizure activity[51]. The conditional deletion of Has2 in other tissues will be necessary to determine its full role in development; given the high HA levels in many developing tissues[60], other critical roles for HAS2 in development will likely be forthcoming.
Alterations in HA synthesis might be expected to affect heart development in humans and this led investigators to screen the DNA from 100 children with a ventricular septal defect for HAS2 mutations[61]. In a single patient, a c.A1496T (p.Glu499Val) mutation was identified that reduced the activity of transiently expressed HAS2 by 40%. This suggests that partial HAS2 deficiency could be the cause of congenital heart defects in other patients. These results have not yet been replicated in independent cohorts.
The HAS3 enzyme is the most active of the HA synthases and produces HA of lower molecular mass[46]. Initial studies of Has3-/- mice showed no obvious phenotype although they were more resistant to ventilator-induced lung injury[62]. However, in recent studies focused on the brain, Has3-/- mice were found to have reduced HA, crowding of neurons and reduced extracellular space in specific regions of the brain[51]. These mice exhibited a significant increase in abnormal electrical activity and a substantial predisposition to seizure activity. The reduced extracellular space in the affected brain regions reduced the diffusion of molecules and provided evidence for a physiological role of HA in the regulation of brain extracellular space. Studies of the role of HAS3 in human epilepsy have not yet been reported and there are only limited data regarding other clinical phenotypes[51].
The first disorder of increased HA synthesis to be described in humans concerned an individual with thickened skin and excessive skin folding[63]. The skin folding was particularly prominent in the face and limbs; skin of the trunk was thickened but not folded. That individual did not have other significant medical issues and the skin folds decreased with age. Histological studies demonstrated excessive extracellular HA in the skin. The concentration of serum HA was markedly increased, up to 1000 fold greater than normal. Although an underlying genetic cause was not reported, the authors speculated that a similar disorder affects the shar-pei dog.
The shar-pei dog was selectively bred to increase the presence and prominence of skin folds, resulting in the meat-mouth shar-pei, which has an extreme presentation of the trait and a condition termed hereditary cutaneous mucinosis. Demonstration that the material accumulating in the skin and serum is HA[64] and that these dogs have elevated levels of HAS2 mRNA[65], led the condition to be renamed as hereditary cutaneous hyaluronosis[66]. Mapping studies linked the severe HA accumulation to duplications upstream of HAS2 and to a familial fever syndrome that is found in shar-pei dogs[67]. However, the linkage between this duplication and the familial fever syndrome has recently been refuted[68].
Another naturally occurring animal model with increased levels of HA in the skin is the naked mole rat, a rodent recognized for its long life span (about 30 years) and resistance to cancer. Investigators seeking to identify the basis of the resistance to cancer discovered that fibroblasts derived from the naked mole rat synthesize very large quantities of high molecular mass HA[69]. The naked mole rat was found to express a reduced level of the HA-degrading enzyme, HYAL2, as well as a unique form of HAS2. HAS2 from the naked mole rat contains substitutions of Ser for Asn at two different and highly conserved regions of the enzyme. Together, these changes in HA metabolism result in increased HA of higher molecular mass. To demonstrate the relationship between the excessive high molecular mass HA and resistance to cancer, the authors used transient expression approaches to alter the levels of HAS2 or HYAL2. Either a reduction in HAS2 by siRNA, or an increase in HYAL2 by transient overexpression, resulted in a decrease in the extracellular HA and susceptibility to tumorigenesis[69].
Thus, in contrast to the severe developmental abnormalities identified in mice with HAS2 deficiency, an overexpression of HAS2 results in a much milder phenotype in rodents and dogs. It is even possible that the excessive synthesis of HA has some protective roles although the studies in the naked mole rat indicated that other factors work in conjunction with HA to mediate the cancer resistance phenotype[69]. In humans, a better understanding of the full range of phenotypes that might be associated with alterations in HAS2 expression awaits the identification of additional patients.
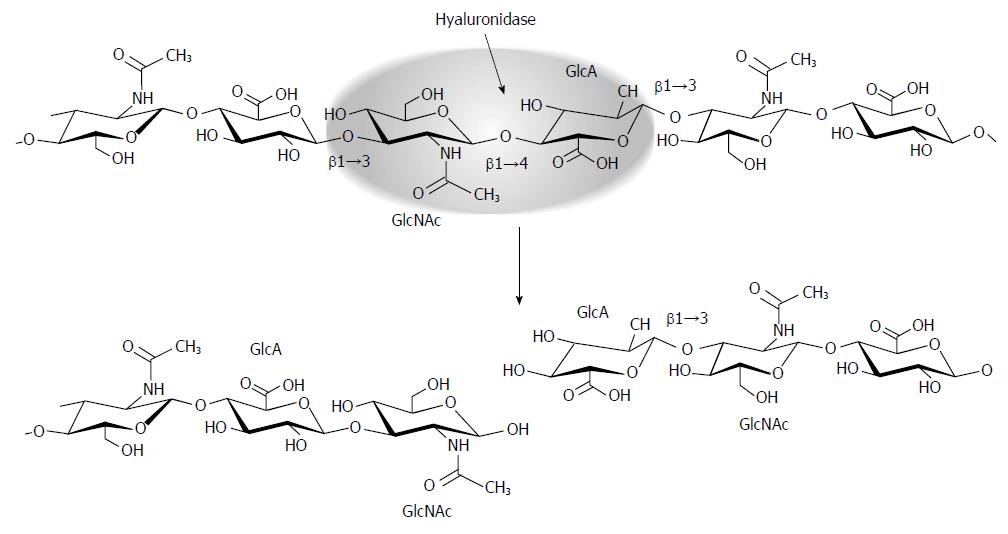
Within the cell, the complete degradation of HA to monosaccharides requires the concerted action of an endoglycosidase, hyaluronidase, and the exoglycosidases β-N-acetyl-D-hexosaminidase and β-D-glucuronidase[70]. As an endoglycosidase, hyaluronidase cleaves the hyaluronan polymer internally at β1→4 linkages between β-N-acetylglucosaminine and β-D-glucuronic acid (Figure 2). The resulting oligosaccharides become substrates for the exoglycosidase β-D-glucuronidase which hydrolyzes the terminal non-reducing glucuronic acid, leaving a non-reducing β-N-acetylglucosaminine that can then be hydrolyzed by β-N-acetyl-D-hexosaminidase[71].
Two forms of hyaluronidase were originally characterized in human tissues, one with a neutral pH optimum that was rich in testes, and a second with an acid pH optimum that was found in most solid tissues and serum[72]. A glycosylphosphatidylinositol (GPI)-linked sperm protein important for fertilization, PH-20, was later found to have sequence similarity to bee venom hyaluronidase[73], and was soon recognized as the neutrally active hyaluronidase[74]; its gene, SPAM1, was mapped to human 7q31 (and mouse chromosome 6A2)[75]. The acid-active hyaluronidase in human plasma, HYAL1, was isolated[76], and mapped to human chromosome 3p21.3 (mouse chromosome 9F1-F2), a region previously characterized as having a tumor suppressor locus[77]. The human genome sequence allowed additional putative hyaluronidases to be identified, including HYAL2 which encodes an enzyme with weak activity toward high molecular mass HA[78] and mapped in close proximity to HYAL1[79]. Bioinformatic approaches revealed two tandemly repeated groups of three genes, which mapped to human chromosome 3p21.3 (centromere-HYAL2-HYAL1-HYAL3-telomere)[80] and 7q31.3 (centromere-SPAM1-HYAL4-HYALP1-telomere)[81,82]. An additional gene, Hyal5, is found downstream of Spam1 in mice[83]. These genes were identified as putative hyaluronidase homologues based on sequence conservation but further studies demonstrated that HYALP1 is an expressed pseudogene in humans[81], HYAL4 encodes a chondroitinase[84,85], and HYAL3 has no detectable in vitro activity[86,87]. Additional expression studies using human and mouse tissues showed that HYAL1/Hyal1, HYAL2/Hyal2, and HYAL3/Hyal3 are broadly expressed in somatic tissues[81,88], while the expression of SPAM1 is limited primarily to the testes[75] and HYAL4 to skeletal muscle and placenta[81]. The recently described HA binding/degradative protein, KIAA1199, is expressed in many tissues including brain, lung, pancreas, testis and ovary but is not expressed in liver, kidney or spleen[89].
The first individual to be identified with a hyaluronidase deficiency was a young girl who had mild short stature, mild facial dysmorphism including a flattened nasal bridge, a bifid uvula and a submucosal cleft palate, and a presenting complaint of multiple periarticular soft tissue masses involving both large and small joints; there was no evidence of visceral or neurological involvement. Histological and ultrastructural analyses of biopsies showed the periarticular masses to have a synovium that was massively expanded with macrophages containing numerous lysosomes containing fibrillar storage material; the skin showed similar pathology with fibroblasts containing increased numbers of lysosomes with excess macromolecular substrate. This work suggested some type of lysosomal storage disorder. Early biochemical investigations disclosed markedly elevated levels of serum HA and a deficiency of acid pH optimum hyaluronidase (and chondroitinase) and the condition, a new lysosomal storage disorder, was termed mucopolysaccharidosis IX[90]. This patient was subsequently found to have a 1361del37ins14 mutation in HYAL1 that was predicted to result in a premature stop codon on one allele, and a c.G1412A (p.Glu268Lys) on the other allele; no HYAL1 activity was detectable[91]. Three additional patients with HYAL1 deficiency have since been identified in a consanguineous family of Saudi Arabian origin[92]. The proband in this family was initially characterized with idiopathic juvenile arthritis that did not respond to treatment with non-steroidal anti-inflammatory drugs. Two siblings of the proband were subsequently found to have joint abnormalities including joint effusions, synovial proliferation, and cysts. No serum HYAL1 activity was detected in any of the patients and all of them were homozygous for a c.104delT (p.Val35AlafsX25) mutation. The phenotypes of all 4 individuals with HYAL1 deficiency are less severe than that predicted for human hyaluronidase deficiency based on the broad distribution of HA in the human body and the significance of HA in many developmental and remodeling processes. Taken together, this information suggests that other enzymes (partially) compensate for the deficiency of HYAL1.
Consistent with the phenotype of persons with mucopolysaccharidosis IX, a Hyal1-/- mouse model did not show a generalized accumulation of HA[93]. The only detectable phenotype was a progressive loss of articular cartilage proteoglycan in the knee joint, starting as early as 3 mo of age. The mild phenotype in the mouse model is likely due, at least in part, to compensation by certain exoglycosidase activities, as mice deficient in both HYAL1 and β-hexosaminidases A and B (Hyal1-/-Hexa-/-Hexb-/-) showed global HA accumulation that was significantly higher than that in either Hyal1-/- or Hexa-/-Hexb-/- mice[94]. It seems likely that except in the joints the levels of β-hexosaminidase and β-glucuronidase are adequate to compensate for HYAL1 deficiency in mice, and by extrapolation, in humans as well. Interestingly, female Hyal1-/- mice had prolonged fertility; the basis for this is currently unclear[95].
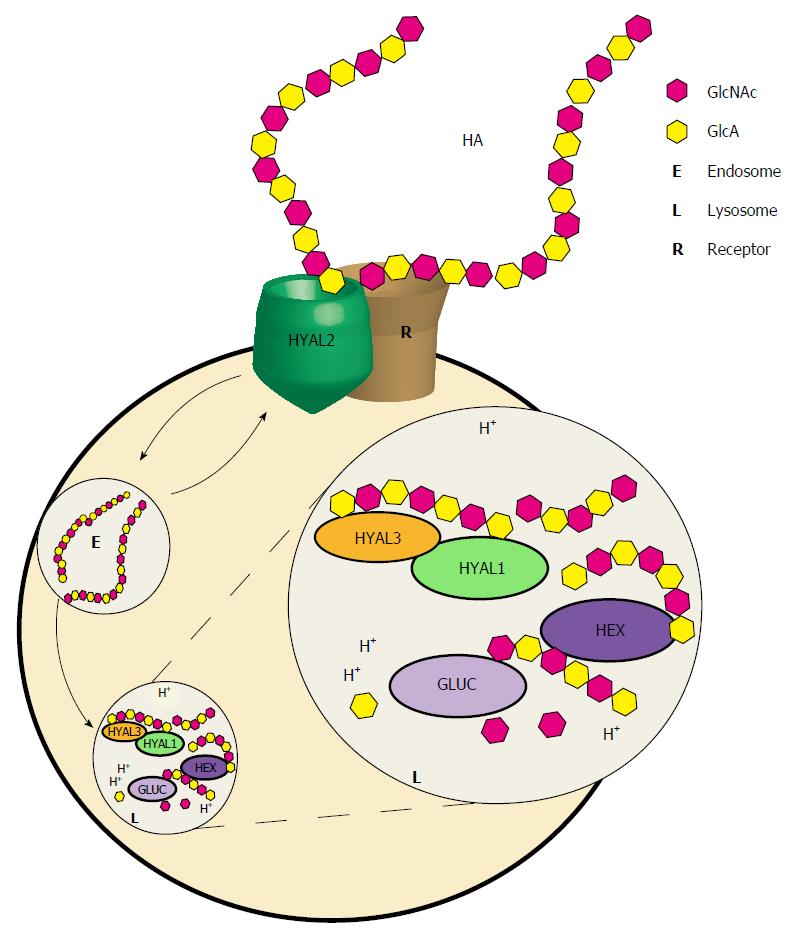
HYAL2 is a broadly expressed glycosylphosphatidylinositol-linked cell surface protein whose localization and activity have been controversial. Its activity is weak, but has been successfully demonstrated toward high molecular mass HA which it appears to cleave to 20 kDa fragments[86,96]. HYAL2 interacts with CD44 and NHE1 proteins[97,98], which has led to the model shown in Figure 3, proposing that HYAL2 initiates the breakdown of extracellular HA, in partnership with the Na/H exchanger isoform 1 (NHE1) which acidifies the local environment[39]. That model further proposes that fragments generated by HYAL2 are internalized by receptor-mediated endocytosis. The generation of a Hyal2-/- mouse which exhibited mild anemia, craniofacial abnormalities, and evidence of pre-weaning lethality, did not completely support this model, as generalized storage of HA was not detected[99]. However, further studies demonstrated substantial (2/3) pre-weaning lethality in Hyal2-/- mice and severe cardiopulmonary dysfunction leading to death in 54% of Hyal2-/- mice by an average of 3.2 mo of age[100]. Accumulating extracellular HA in the heart, lung, and serum of these mice was of higher than average molecular mass, consistent with the model that HYAL2 may be important in initiating the depolymerization of HA in the extracellular matrix. The pre-weaning lethality in the mice suggests that a complete deficiency of HYAL2 may cause a severe, if not lethal, condition in humans, although the genetic background may provide important modifying effects that might either mitigate or exacerbate any of the above mentioned monogenic biosynthetic or catabolic enzyme disorders.
HYAL3 is broadly but weakly expressed in somatic cells. Although it is homologous to the other hyaluronidases, no activity has been clearly associated with the enzyme[86,87]. Hyal3-/- mice did not accumulate HA, and the only detectable phenotype was a minor change in the histopathology of the lungs[101]. However, the overexpression of HYAL3 in cultured baby hamster kidney cells resulted in increased HYAL1 activity, suggesting a role for HYAL3 in HA metabolism[87].
PH-20 (SPAM1) plays a role in both penetration of the cumulus barrier and binding of the zona pellucida during fertilization[102,103]. When it was initially mapped to human chromosome 7q31 and shown to be specifically expressed in the testes, it was thought that mutations in the gene might be identified as a cause of infertility[75], but no reports of mutations have been made. Studies in Spam1-/- mice have not provided insights regarding phenotype(s) that might be associated with human SPAM1 mutations because HYAL5 in mice can compensate for SPAM1 deficiency[104-106].
It is clear that the range of conditions that result from abnormalities in HA metabolizing enzymes is only partially understood. Monogenic dysfunction of enzymes involved in HA synthesis and HA catabolism can have marked phenotypic effects that can involve many tissues and organ systems. It is likely that the ongoing broad application of next generation sequencing to identify the causes of unexplained phenotypes in humans will identify novel human conditions that result from variations in the HYAL and HAS enzymes and broaden the spectrum of already described conditions, furthering our understanding of their in vivo functions.
P- Reviewer: Frade JM, Hegardt FG, Scatena R S- Editor: Tian YL L- Editor: A E- Editor: Wang CH
| 2. | Itano N, Kimata K. Mammalian hyaluronan synthases. IUBMB Life. 2002;54:195-199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 300] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 3. | Laurent TC, Fraser JRE. Catabolism of hyaluronan. Degradation of Bioactive Substances: Physiology and Pathology. Boca Raton: CRC Press 1991; 249-265. [Cited in This Article: ] |
| 4. | Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med. 1997;242:27-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1386] [Cited by in F6Publishing: 1299] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 5. | Toole BP. Hyaluronan in morphogenesis. J Intern Med. 1997;242:35-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 261] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 6. | Engström-Laurent A, Laurent UB, Lilja K, Laurent TC. Concentration of sodium hyaluronate in serum. Scand J Clin Lab Invest. 1985;45:497-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 191] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 7. | Delpech B, Bertrand P, Maingonnat C. Immunoenzymoassay of the hyaluronic acid-hyaluronectin interaction: application to the detection of hyaluronic acid in serum of normal subjects and cancer patients. Anal Biochem. 1985;149:555-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 90] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Laurent TC, Laurent UBG, Fraser JRE. Serum hyaluronan as a disease marker. Ann Med. 1996;28:241-253. [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 123] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 9. | Reed RK, Lilja K, Laurent TC. Hyaluronan in the rat with special reference to the skin. Acta Physiol Scand. 1988;134:405-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 122] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Petrey AC, de la Motte CA. Hyaluronan, a crucial regulator of inflammation. Front Immunol. 2014;5:101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 317] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 11. | Salustri A, Camaioni A, Di Giacomo M, Fulop C, Hascall VC. Hyaluronan and proteoglycans in ovarian follicles. Hum Reprod Update. 1999;5:293-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 89] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Stern R, Asari AA, Sugahara KN. Hyaluronan fragments: an information-rich system. Eur J Cell Biol. 2006;85:699-715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 787] [Cited by in F6Publishing: 787] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 13. | Erickson M, Stern R. Chain gangs: new aspects of hyaluronan metabolism. Biochem Res Int. 2012;2012:893947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Schiller S, Mathews MB, Goldfaber L, Ludowieg J, Dorfman A. The metabolism of mucopolysaccharides in animals. II. Studies in skin utilizing labeled acetate. J Biol Chem. 1955;212:531-535. [PubMed] [Cited in This Article: ] |
| 15. | Schiller S, Mathews MB, Cifonelli JA, Dorfman A. The metabolism of mucopolysaccharides in animals. III. Further studies on skin utilizing C14-glucose, C14-acetate, and S35-sodium sulfate. J Biol Chem. 1956;218:139-145. [PubMed] [Cited in This Article: ] |
| 16. | Fraser JR, Laurent TC, Pertoft H, Baxter E. Plasma clearance, tissue distribution and metabolism of hyaluronic acid injected intravenously in the rabbit. Biochem J. 1981;200:415-424. [PubMed] [Cited in This Article: ] |
| 17. | Fraser JR, Laurent TC, Engström-Laurent A, Laurent UG. Elimination of hyaluronic acid from the blood stream in the human. Clin Exp Pharmacol Physiol. 1984;11:17-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 143] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 18. | Schiller S, Dorfman A. The metabolism of mucopolysaccharides in animals. IV. The influence of insulin. J Biol Chem. 1957;227:625-632. [PubMed] [Cited in This Article: ] |
| 19. | Hascall V, Sandy JD, Handley CJ. In: Archer CW, Caterson B, editors. Regulation of proteoglycan metabolism in articular cartilage. Biology of the Synovial Joint. Amsterdam: Harwood Academic Publishers 1999; 101-120. [Cited in This Article: ] |
| 20. | Fraser JR, Laurent TC. Turnover and metabolism of hyaluronan. Ciba Found Symp. 1989;143:41-53; discussion 53-59, 281-285. [PubMed] [Cited in This Article: ] |
| 21. | Fraser JR, Appelgren LE, Laurent TC. Tissue uptake of circulating hyaluronic acid. A whole body autoradiographic study. Cell Tissue Res. 1983;233:285-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 80] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Eriksson S, Fraser JR, Laurent TC, Pertoft H, Smedsrød B. Endothelial cells are a site of uptake and degradation of hyaluronic acid in the liver. Exp Cell Res. 1983;144:223-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 210] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 23. | Fraser JR, Alcorn D, Laurent TC, Robinson AD, Ryan GB. Uptake of circulating hyaluronic acid by the rat liver. Cellular localization in situ. Cell Tissue Res. 1985;242:505-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 108] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Smedsrød B, Pertoft H, Eriksson S, Fraser JR, Laurent TC. Studies in vitro on the uptake and degradation of sodium hyaluronate in rat liver endothelial cells. Biochem J. 1984;223:617-626. [PubMed] [Cited in This Article: ] |
| 25. | Laurent TC, Fraser JR, Pertoft H, Smedsrød B. Binding of hyaluronate and chondroitin sulphate to liver endothelial cells. Biochem J. 1986;234:653-658. [PubMed] [Cited in This Article: ] |
| 26. | Laurent TC, Lilja K, Brunnberg L, Engström-Laurent A, Laurent UB, Lindqvist U, Murata K, Ytterberg D. Urinary excretion of hyaluronan in man. Scand J Clin Lab Invest. 1987;47:793-799. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 53] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Falkowski M, Schledzewski K, Hansen B, Goerdt S. Expression of stabilin-2, a novel fasciclin-like hyaluronan receptor protein, in murine sinusoidal endothelia, avascular tissues, and at solid/liquid interfaces. Histochem Cell Biol. 2003;120:361-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | McGary CT, Raja RH, Weigel PH. Endocytosis of hyaluronic acid by rat liver endothelial cells. Evidence for receptor recycling. Biochem J. 1989;257:875-884. [PubMed] [Cited in This Article: ] |
| 29. | Weigel JA, Raymond RC, McGary C, Singh A, Weigel PH. A blocking antibody to the hyaluronan receptor for endocytosis (HARE) inhibits hyaluronan clearance by perfused liver. J Biol Chem. 2003;278:9808-9812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 69] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Hirose Y, Saijou E, Sugano Y, Takeshita F, Nishimura S, Nonaka H, Chen YR, Sekine K, Kido T, Nakamura T. Inhibition of Stabilin-2 elevates circulating hyaluronic acid levels and prevents tumor metastasis. Proc Natl Acad Sci USA. 2012;109:4263-4268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 31. | Culty M, Nguyen HA, Underhill CB. The hyaluronan receptor (CD44) participates in the uptake and degradation of hyaluronan. J Cell Biol. 1992;116:1055-1062. [PubMed] [Cited in This Article: ] |
| 32. | Prevo R, Banerji S, Ferguson DJ, Clasper S, Jackson DG. Mouse LYVE-1 is an endocytic receptor for hyaluronan in lymphatic endothelium. J Biol Chem. 2001;276:19420-19430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 351] [Cited by in F6Publishing: 346] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 33. | Nightingale TD, Frayne ME, Clasper S, Banerji S, Jackson DG. A mechanism of sialylation functionally silences the hyaluronan receptor LYVE-1 in lymphatic endothelium. J Biol Chem. 2009;284:3935-3945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Luong MX, Tam J, Lin Q, Hagendoorn J, Moore KJ, Padera TP, Seed B, Fukumura D, Kucherlapati R, Jain RK. Lack of lymphatic vessel phenotype in LYVE-1/CD44 double knockout mice. J Cell Physiol. 2009;219:430-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Wang A, de la Motte C, Lauer M, Hascall V. Hyaluronan matrices in pathobiological processes. FEBS J. 2011;278:1412-1418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 36. | Hua Q, Knudson CB, Knudson W. Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J Cell Sci. 1993;106:365-375. [PubMed] [Cited in This Article: ] |
| 37. | Wheatley SC, Isacke CM, Crossley PH. Restricted expression of the hyaluronan receptor, CD44, during postimplantation mouse embryogenesis suggests key roles in tissue formation and patterning. Development. 1993;119:295-306. [PubMed] [Cited in This Article: ] |
| 38. | Fraser JR, Kimpton WG, Laurent TC, Cahill RN, Vakakis N. Uptake and degradation of hyaluronan in lymphatic tissue. Biochem J. 1988;256:153-158. [PubMed] [Cited in This Article: ] |
| 39. | Stern R. Devising a pathway for hyaluronan catabolism: are we there yet? Glycobiology. 2003;13:105R-115R. [PubMed] [Cited in This Article: ] |
| 40. | Ng CK, Handley CJ, Preston BN, Robinson HC. The extracellular processing and catabolism of hyaluronan in cultured adult articular cartilage explants. Arch Biochem Biophys. 1992;298:70-79. [PubMed] [Cited in This Article: ] |
| 41. | Yoshida H, Nagaoka A, Kusaka-Kikushima A, Tobiishi M, Kawabata K, Sayo T, Sakai S, Sugiyama Y, Enomoto H, Okada Y. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci USA. 2013;110:5612-5617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 198] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 42. | Abe S, Katagiri T, Saito-Hisaminato A, Usami S, Inoue Y, Tsunoda T, Nakamura Y. Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am J Hum Genet. 2003;72:73-82. [PubMed] [Cited in This Article: ] |
| 43. | Yoshida H, Nagaoka A, Nakamura S, Sugiyama Y, Okada Y, Inoue S. Murine homologue of the human KIAA1199 is implicated in hyaluronan binding and depolymerization. FEBS Open Bio. 2013;3:352-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 44. | Weigel PH, DeAngelis PL. Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J Biol Chem. 2007;282:36777-36781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 250] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 45. | Spicer AP, Seldin MF, Olsen AS, Brown N, Wells DE, Doggett NA, Itano N, Kimata K, Inazawa J, McDonald JA. Chromosomal localization of the human and mouse hyaluronan synthase genes. Genomics. 1997;41:493-497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 82] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, Shinomura T, Hamaguchi M, Yoshida Y, Ohnuki Y. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J Biol Chem. 1999;274:25085-25092. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 650] [Cited by in F6Publishing: 653] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 47. | Rilla K, Oikari S, Jokela TA, Hyttinen JM, Kärnä R, Tammi RH, Tammi MI. Hyaluronan synthase 1 (HAS1) requires higher cellular UDP-GlcNAc concentration than HAS2 and HAS3. J Biol Chem. 2013;288:5973-5983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 48. | Törrönen K, Nikunen K, Kärnä R, Tammi M, Tammi R, Rilla K. Tissue distribution and subcellular localization of hyaluronan synthase isoenzymes. Histochem Cell Biol. 2014;141:17-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 49. | Spicer AP, McDonald JA. Characterization and molecular evolution of a vertebrate hyaluronan synthase gene family. J Biol Chem. 1998;273:1923-1932. [PubMed] [Cited in This Article: ] |
| 50. | Spicer AP, Tien JL, Joo A, Bowling RA. Investigation of hyaluronan function in the mouse through targeted mutagenesis. Glycoconj J. 2002;19:341-345. [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 51. | Arranz AM, Perkins KL, Irie F, Lewis DP, Hrabe J, Xiao F, Itano N, Kimata K, Hrabetova S, Yamaguchi Y. Hyaluronan deficiency due to Has3 knock-out causes altered neuronal activity and seizures via reduction in brain extracellular space. J Neurosci. 2014;34:6164-6176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 103] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 52. | Mack JA, Feldman RJ, Itano N, Kimata K, Lauer M, Hascall VC, Maytin EV. Enhanced inflammation and accelerated wound closure following tetraphorbol ester application or full-thickness wounding in mice lacking hyaluronan synthases Has1 and Has3. J Invest Dermatol. 2012;132:198-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 53. | Adamia S, Crainie M, Kriangkum J, Mant MJ, Belch AR, Pilarski LM. Abnormal expression of hyaluronan synthases in patients with Waldenstrom’s macroglobulimenia. Semin Oncol. 2003;30:165-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 54. | Adamia S, Reiman T, Crainie M, Mant MJ, Belch AR, Pilarski LM. Intronic splicing of hyaluronan synthase 1 (HAS1): a biologically relevant indicator of poor outcome in multiple myeloma. Blood. 2005;105:4836-4844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 52] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Kuppusamy H, Ogmundsdottir HM, Baigorri E, Warkentin A, Steingrimsdottir H, Haraldsdottir V, Mant MJ, Mackey J, Johnston JB, Adamia S. Inherited polymorphisms in hyaluronan synthase 1 predict risk of systemic B-cell malignancies but not of breast cancer. PLoS One. 2014;9:e100691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 56. | Adamia S, Reichert AA, Kuppusamy H, Kriangkum J, Ghosh A, Hodges JJ, Pilarski PM, Treon SP, Mant MJ, Reiman T. Inherited and acquired variations in the hyaluronan synthase 1 (HAS1) gene may contribute to disease progression in multiple myeloma and Waldenstrom macroglobulinemia. Blood. 2008;112:5111-5121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 57. | Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349-360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 711] [Cited by in F6Publishing: 646] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 58. | Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med. 2002;8:850-855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 239] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 59. | Matsumoto K, Li Y, Jakuba C, Sugiyama Y, Sayo T, Okuno M, Dealy CN, Toole BP, Takeda J, Yamaguchi Y. Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development. 2009;136:2825-2835. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 60. | Fenderson BA, Stamenkovic I, Aruffo A. Localization of hyaluronan in mouse embryos during implantation, gastrulation and organogenesis. Differentiation. 1993;54:85-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 44] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 61. | Zhu X, Deng X, Huang G, Wang J, Yang J, Chen S, Ma X, Wang B. A novel mutation of Hyaluronan synthase 2 gene in Chinese children with ventricular septal defect. PLoS One. 2014;9:e87437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 62. | Bai KJ, Spicer AP, Mascarenhas MM, Yu L, Ochoa CD, Garg HG, Quinn DA. The role of hyaluronan synthase 3 in ventilator-induced lung injury. Am J Respir Crit Care Med. 2005;172:92-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 63. | Ramsden CA, Bankier A, Brown TJ, Cowen PS, Frost GI, McCallum DD, Studdert VP, Fraser JR. A new disorder of hyaluronan metabolism associated with generalized folding and thickening of the skin. J Pediatr. 2000;136:62-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 64. | Zanna G, Fondevila D, Bardagí M, Docampo MJ, Bassols A, Ferrer L. Cutaneous mucinosis in shar-pei dogs is due to hyaluronic acid deposition and is associated with high levels of hyaluronic acid in serum. Vet Dermatol. 2008;19:314-318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 65. | Zanna G, Docampo MJ, Fondevila D, Bardagí M, Bassols A, Ferrer L. Hereditary cutaneous mucinosis in shar pei dogs is associated with increased hyaluronan synthase-2 mRNA transcription by cultured dermal fibroblasts. Vet Dermatol. 2009;20:377-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 66. | Docampo MJ, Zanna G, Fondevila D, Cabrera J, López-Iglesias C, Carvalho A, Cerrato S, Ferrer L, Bassols A. Increased HAS2-driven hyaluronic acid synthesis in shar-pei dogs with hereditary cutaneous hyaluronosis (mucinosis). Vet Dermatol. 2011;22:535-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 67. | Olsson M, Meadows JR, Truvé K, Rosengren Pielberg G, Puppo F, Mauceli E, Quilez J, Tonomura N, Zanna G, Docampo MJ. A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs. PLoS Genet. 2011;7:e1001332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 68. | Metzger J, Distl O. A study of Shar-Pei dogs refutes association of the ‘meatmouth’ duplication near HAS2 with Familial Shar-Pei Fever. Anim Genet. 2014;45:763-764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 69. | Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, Mao Z, Nevo E, Gorbunova V, Seluanov A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 501] [Cited by in F6Publishing: 507] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 70. | Rodén L, Campbell P, Fraser JR, Laurent TC, Pertoft H, Thompson JN. Enzymic pathways of hyaluronan catabolism. Ciba Found Symp. 1989;143:60-76; discussion 76-86, 281-285. [PubMed] [Cited in This Article: ] |
| 71. | Linker A, Meyer K, Weissmann B. Enzumatic formation of monosaccharides from hyaluronate. J Biol Chem. 1955;213:237-248. [PubMed] [Cited in This Article: ] |
| 72. | De Salegui M, Plonska H, Pigman W. A comparison of serum and testicular hyaluronidase. Arch Biochem Biophys. 1967;121:548-554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 73. | Gmachl M, Kreil G. Bee venom hyaluronidase is homologous to a membrane protein of mammalian sperm. Proc Natl Acad Sci USA. 1993;90:3569-3573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 139] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 74. | Gmachl M, Sagan S, Ketter S, Kreil G. The human sperm protein PH-20 has hyaluronidase activity. FEBS Lett. 1993;336:545-548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 107] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 75. | Jones MH, Davey PM, Aplin H, Affara NA. Expression analysis, genomic structure, and mapping to 7q31 of the human sperm adhesion molecule gene SPAM1. Genomics. 1995;29:796-800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 76. | Frost GI, Csóka AB, Wong T, Stern R. Purification, cloning, and expression of human plasma hyaluronidase. Biochem Biophys Res Commun. 1997;236:10-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 182] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 77. | Csóka AB, Frost GI, Heng HH, Scherer SW, Mohapatra G, Stern R. The hyaluronidase gene HYAL1 maps to chromosome 3p21.2-p21.3 in human and 9F1-F2 in mouse, a conserved candidate tumor suppressor locus. Genomics. 1998;48:63-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 78. | Lepperdinger G, Strobl B, Kreil G. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J Biol Chem. 1998;273:22466-22470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 232] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 79. | Strobl B, Wechselberger C, Beier DR, Lepperdinger G. Structural organization and chromosomal localization of Hyal2, a gene encoding a lysosomal hyaluronidase. Genomics. 1998;53:214-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 80. | Wei MH, Latif F, Bader S, Kashuba V, Chen JY, Duh FM, Sekido Y, Lee CC, Geil L, Kuzmin I. Construction of a 600-kilobase cosmid clone contig and generation of a transcriptional map surrounding the lung cancer tumor suppressor gene (TSG) locus on human chromosome 3p21.3: progress toward the isolation of a lung cancer TSG. Cancer Res. 1996;56:1487-1492. [PubMed] [Cited in This Article: ] |
| 81. | Csóka AB, Scherer SW, Stern R. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics. 1999;60:356-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 184] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 82. | Csoka AB, Frost GI, Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 431] [Cited by in F6Publishing: 435] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 83. | Kim E, Baba D, Kimura M, Yamashita M, Kashiwabara S, Baba T. Identification of a hyaluronidase, Hyal5, involved in penetration of mouse sperm through cumulus mass. Proc Natl Acad Sci USA. 2005;102:18028-18033. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 84. | Chatel A, Hemming R, Hobert J, Natowicz MR, Triggs-Raine B, Merz DC. The C. elegans hyaluronidase: a developmentally significant enzyme with chondroitin-degrading activity at both acidic and neutral pH. Matrix Biol. 2010;29:494-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 85. | Kaneiwa T, Mizumoto S, Sugahara K, Yamada S. Identification of human hyaluronidase-4 as a novel chondroitin sulfate hydrolase that preferentially cleaves the galactosaminidic linkage in the trisulfated tetrasaccharide sequence. Glycobiology. 2010;20:300-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 86. | Harada H, Takahashi M. CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J Biol Chem. 2007;282:5597-5607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 240] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 87. | Hemming R, Martin DC, Slominski E, Nagy JI, Halayko AJ, Pind S, Triggs-Raine B. Mouse Hyal3 encodes a 45- to 56-kDa glycoprotein whose overexpression increases hyaluronidase 1 activity in cultured cells. Glycobiology. 2008;18:280-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 88. | Shuttleworth TL, Wilson MD, Wicklow BA, Wilkins JA, Triggs-Raine BL. Characterization of the murine hyaluronidase gene region reveals complex organization and cotranscription of Hyal1 with downstream genes, Fus2 and Hyal3. J Biol Chem. 2002;277:23008-23018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 89. | Michishita E, Garcés G, Barrett JC, Horikawa I. Upregulation of the KIAA1199 gene is associated with cellular mortality. Cancer Lett. 2006;239:71-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 90. | Natowicz MR, Short MP, Wang Y, Dickersin GR, Gebhardt MC, Rosenthal DI, Sims KB, Rosenberg AE. Clinical and biochemical manifestations of hyaluronidase deficiency. N Engl J Med. 1996;335:1029-1033. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 113] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 91. | Triggs-Raine B, Salo TJ, Zhang H, Wicklow BA, Natowicz MR. Mutations in HYAL1, a member of a tandemly distributed multigene family encoding disparate hyaluronidase activities, cause a newly described lysosomal disorder, mucopolysaccharidosis IX. Proc Natl Acad Sci USA. 1999;96:6296-6300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 134] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 92. | Imundo L, Leduc CA, Guha S, Brown M, Perino G, Gushulak L, Triggs-Raine B, Chung WK. A complete deficiency of Hyaluronoglucosaminidase 1 (HYAL1) presenting as familial juvenile idiopathic arthritis. J Inherit Metab Dis. 2011;34:1013-1022. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 93. | Martin DC, Atmuri V, Hemming RJ, Farley J, Mort JS, Byers S, Hombach-Klonisch S, Csoka AB, Stern R, Triggs-Raine BL. A mouse model of human mucopolysaccharidosis IX exhibits osteoarthritis. Hum Mol Genet. 2008;17:1904-1915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 94. | Gushulak L, Hemming R, Martin D, Seyrantepe V, Pshezhetsky A, Triggs-Raine B. Hyaluronidase 1 and β-hexosaminidase have redundant functions in hyaluronan and chondroitin sulfate degradation. J Biol Chem. 2012;287:16689-16697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 95. | Dumaresq-Doiron K, Edjekouane L, Orimoto AM, Yoffou PH, Gushulak L, Triggs-Raine B, Carmona E. Hyal-1 but not Hyal-3 deficiency has an impact on ovarian folliculogenesis and female fertility by altering the follistatin/activin/Smad3 pathway and the apoptotic process. J Cell Physiol. 2012;227:1911-1922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 96. | Lepperdinger G, Müllegger J, Kreil G. Hyal2--less active, but more versatile? Matrix Biol. 2001;20:509-514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 123] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 97. | Duterme C, Mertens-Strijthagen J, Tammi M, Flamion B. Two novel functions of hyaluronidase-2 (Hyal2) are formation of the glycocalyx and control of CD44-ERM interactions. J Biol Chem. 2009;284:33495-33508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 98. | Bourguignon LY, Singleton PA, Diedrich F, Stern R, Gilad E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J Biol Chem. 2004;279:26991-27007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 312] [Cited by in F6Publishing: 319] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 99. | Jadin L, Wu X, Ding H, Frost GI, Onclinx C, Triggs-Raine B, Flamion B. Skeletal and hematological anomalies in HYAL2-deficient mice: a second type of mucopolysaccharidosis IX? FASEB J. 2008;22:4316-4326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 100. | Chowdhury B, Hemming R, Hombach-Klonisch S, Flamion B, Triggs-Raine B. Murine hyaluronidase 2 deficiency results in extracellular hyaluronan accumulation and severe cardiopulmonary dysfunction. J Biol Chem. 2013;288:520-528. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 101. | Atmuri V, Martin DC, Hemming R, Gutsol A, Byers S, Sahebjam S, Thliveris JA, Mort JS, Carmona E, Anderson JE. Hyaluronidase 3 (HYAL3) knockout mice do not display evidence of hyaluronan accumulation. Matrix Biol. 2008;27:653-660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 102. | Lin Y, Mahan K, Lathrop WF, Myles DG, Primakoff P. A hyaluronidase activity of the sperm plasma membrane protein PH-20 enables sperm to penetrate the cumulus cell layer surrounding the egg. J Cell Biol. 1994;125:1157-1163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 196] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 103. | Sabeur K, Cherr GN, Yudin AI, Primakoff P, Li MW, Overstreet JW. The PH-20 protein in human spermatozoa. J Androl. 1997;18:151-158. [PubMed] [Cited in This Article: ] |
| 104. | Baba D, Kashiwabara S, Honda A, Yamagata K, Wu Q, Ikawa M, Okabe M, Baba T. Mouse sperm lacking cell surface hyaluronidase PH-20 can pass through the layer of cumulus cells and fertilize the egg. J Biol Chem. 2002;277:30310-30314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 137] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 105. | Reitinger S, Laschober GT, Fehrer C, Greiderer B, Lepperdinger G. Mouse testicular hyaluronidase-like proteins SPAM1 and HYAL5 but not HYALP1 degrade hyaluronan. Biochem J. 2007;401:79-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 106. | Kimura M, Kim E, Kang W, Yamashita M, Saigo M, Yamazaki T, Nakanishi T, Kashiwabara S, Baba T. Functional roles of mouse sperm hyaluronidases, HYAL5 and SPAM1, in fertilization. Biol Reprod. 2009;81:939-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 72] [Article Influence: 4.8] [Reference Citation Analysis (0)] |